













| General Information: |
|
| Name(s) found: |
PANK2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 901 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGQEDEYDP ILDNKREAEA KSQVSVAADK NMAPSTSGTP IHRSGSRPQL DLSKAEIQGN 60
61 LEERDPTILL PNQSDDISHL ALDIGGSLIK LLYFSRHEDY SNDDDKRKRT IKERLGITNG 120
121 NLRSYPVLGG RLHFVKFETH KINECLDFIH SKQLHRRDPY PWSSKTLPLG TGVIKVTGGG 180
181 AFKFADLFKE RLGVSIEKED EMHCLVSGAN FLLKAIRHEA FTHMEGEKEF VQIDPNDLYP 240
241 YLLVNVGSGV SIIKVDGEGK FERVSGTNVG GGTYWGLGRL LTKCKSFDEL LELSQKGDNS 300
301 AIDMLVGDIY GGMDYSKIGL SASTIASSFG KAISENKELD DYRPEDISLS LLRMISYNIG 360
361 QISYLNALRF GLKRIFFGGF FIRGHAYTMD TISFAVHFWS KGEMQAMFLR HEGFLGALGA 420
421 FMSYEKHGLD DLMSHQLVER FPMGAPYTGG NIHGPPLGDL DEKISWMEKF VRRGTEITAP 480
481 VPMTPSKTTG LGGFEVPSSR GSALRSDASA LNVGVLHLVP TLEVFPLLAD PKTYEPNTID 540
541 LSDQGEREYW LKVLSEHLPD LVDTAVASEG GTEDAKRRGD AFARAFSAHL ARLMEEPAAY 600
601 GKLGLANLLE LREECLREFQ FVDAYRSIKQ RENEASLAVL PDLLEELDSM SEEARLLTLI 660
661 EGVLAANIFD WGSRACVDLY HKGTIIEIYR MSRNKMQRPW RVDDFDAFKE RMLGSGGKQP 720
721 HRHKRALLFV DNSGADVILG MLPLAREFLR RGTEVVLVAN SLPALNDVTA MELPDIVAGA 780
781 AKHCDILRRA AEMGGLLVDA MVNPGDGSKK DSTSAPLMVV ENGCGSPCID LRQVSSELAA 840
841 AAKDADLVVL EGMGRALHTN FNAQFQCEAL KLAMVKNQRL AEKLIKGNIY DCVCRYEPPS 900
901 L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
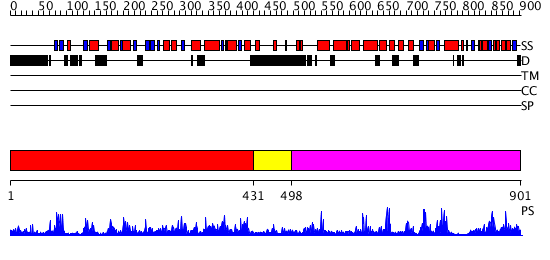
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..430] | 117.0 | No description for 2i7nA was found. | |
| 2 | View Details | [431..497] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [498..901] | 78.154902 | X-ray structure of gene product from Arabidopsis thaliana At2g17340 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)