













| General Information: |
|
| Name(s) found: |
ANL2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 802 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
root development
[IMP]
anthocyanin accumulation in tissues in response to UV light [IMP] |
| Molecular Function: |
transcription factor activity
[ISS]
transcription regulator activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNFGSLFDNT PGGGSTGARL LSGLSYGNHT AATNVLPGGA MAQAAAAASL FSPPLTKSVY 60
61 ASSGLSLALE QPERGTNRGE ASMRNNNNVG GGGDTFDGSV NRRSREEEHE SRSGSDNVEG 120
121 ISGEDQDAAD KPPRKKRYHR HTPQQIQELE SMFKECPHPD EKQRLELSKR LCLETRQVKF 180
181 WFQNRRTQMK TQLERHENAL LRQENDKLRA ENMSIREAMR NPICTNCGGP AMLGDVSLEE 240
241 HHLRIENARL KDELDRVCNL TGKFLGHHHN HHYNSSLELA VGTNNNGGHF AFPPDFGGGG 300
301 GCLPPQQQQS TVINGIDQKS VLLELALTAM DELVKLAQSE EPLWVKSLDG ERDELNQDEY 360
361 MRTFSSTKPT GLATEASRTS GMVIINSLAL VETLMDSNRW TEMFPCNVAR ATTTDVISGG 420
421 MAGTINGALQ LMNAELQVLS PLVPVRNVNF LRFCKQHAEG VWAVVDVSID PVRENSGGAP 480
481 VIRRLPSGCV VQDVSNGYSK VTWVEHAEYD ENQIHQLYRP LLRSGLGFGS QRWLATLQRQ 540
541 CECLAILISS SVTSHDNTSI TPGGRKSMLK LAQRMTFNFC SGISAPSVHN WSKLTVGNVD 600
601 PDVRVMTRKS VDDPGEPPGI VLSAATSVWL PAAPQRLYDF LRNERMRCEW DILSNGGPMQ 660
661 EMAHITKGQD QGVSLLRSNA MNANQSSMLI LQETCIDASG ALVVYAPVDI PAMHVVMNGG 720
721 DSSYVALLPS GFAVLPDGGI DGGGSGDGDQ RPVGGGSLLT VAFQILVNNL PTAKLTVESV 780
781 ETVNNLISCT VQKIRAALQC ES |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
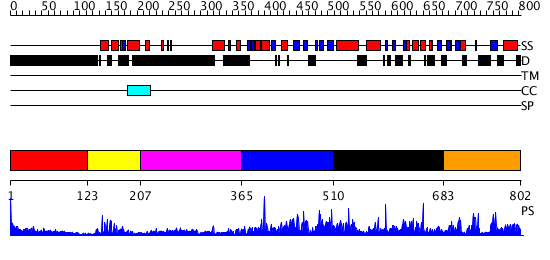
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..122] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [123..206] | 15.69897 | No description for 2r5yA was found. | |
| 3 | View Details | [207..364] | 3.743997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [365..509] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [510..682] | 1.067962 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [683..802] | 2.066976 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)