













| General Information: |
|
| Name(s) found: |
gi|42566965
[NCBI NR]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 859 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATLKPSFTV SPPNSNPIRF GSFPPQCFLR VPKPRRLILP RFRKTTGGGG GLIRCELPDF 60
61 HLSATATTVS GVSTVNLVDQ VQIPKGEMWS VHKFGGTCVG NSQRIRNVAE VIINDNSERK 120
121 LVVVSAMSKV TDMMYDLIRK AQSRDDSYLS ALEAVLEKHR LTARDLLDGD DLASFLSHLH 180
181 NDISNLKAML RAIYIAGHAS ESFSDFVAGH GELWSAQMLS YVVRKTGLEC KWMDTRDVLI 240
241 VNPTSSNQVD PDFGESEKRL DKWFSLNPSK IIIATGFIAS TPQNIPTTLK RDGSDFSAAI 300
301 MGALLRARQV TIWTDVDGVY SADPRKVNEA VILQTLSYQE AWEMSYFGAN VLHPRTIIPV 360
361 MRYNIPIVIR NIFNLSAPGT IICQPPEDDY DLKLTTPVKG FATIDNLALI NVEGTGMAGV 420
421 PGTASDIFGC VKDVGANVIM ISQASSEHSV CFAVPEKEVN AVSEALRSRF SEALQAGRLS 480
481 QIEVIPNCSI LAAVGQKMAS TPGVSCTLFS ALAKANINVR AISQGCSEYN VTVVIKREDS 540
541 VKALRAVHSR FFLSRTTLAM GIVGPGLIGA TLLDQLRDQA AVLKQEFNID LRVLGITGSK 600
601 KMLLSDIGID LSRWRELLNE KGTEADLDKF TQQVHGNHFI PNSVVVDCTA DSAIASRYYD 660
661 WLRKGIHVIT PNKKANSGPL DQYLKLRDLQ RKSYTHYFYE ATVGAGLPII STLRGLLETG 720
721 DKILRIEGIC SGTLSYLFNN FVGDRSFSEV VTEAKNAGFT EPDPRDDLSG TDVARKVIIL 780
781 ARESGLKLDL ADLPIRSLVP EPLKGCTSVE EFMEKLPQYD GDLAKERLDA ENSGEVRLFT 840
841 TNVFPFDQCD HILTIYICM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
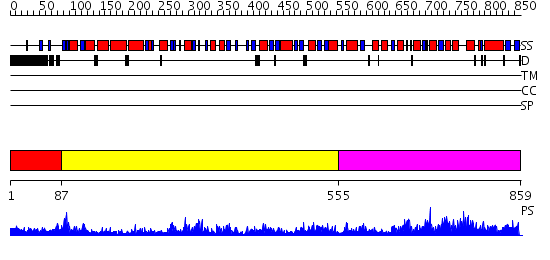
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..86] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [87..554] | 97.522879 | No description for 2hmfA was found. | |
| 3 | View Details | [555..859] | 66.221849 | Homoserine Dehydrogenase in complex with suicide inhibitor complex NAD-5-hydroxy-4-Oxonorvaline |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)