













| General Information: |
|
| Name(s) found: |
GYRA_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 950 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleoid
[IDA]
chloroplast [IDA] mitochondrion [IDA] |
| Biological Process: |
DNA topological change
[IEA]
DNA metabolic process [IEA] |
| Molecular Function: |
DNA binding
[IEA]
ATP binding [IEA][ISS] catalytic activity [ISS] DNA topoisomerase activity [IEA] DNA topoisomerase (ATP-hydrolyzing) activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTPVLCHSTA SIPNPNSLMS LSSTLRLSSS LLRRSFFRFP LTDPLCRLRR TEPSATRFFS 60
61 SRTPRSGKFV VGAGKRGDEQ VKEESGANNG GLVVSGDESR IVPFELHKEA TESYMSYALS 120
121 VLLGRALPDV RDGLKPVHRR ILFAMHELGM SSKKPYKKCA RVVGEVLGKF HPHGDTAVYD 180
181 SLVRMAQSFS LRCPLIQGHG NFGSIDADPP AAMRYTECRL DPLAEAVLLS DLDQDTVDFV 240
241 ANFDNSQKEP AVLPARLPAL LLNGASGIAV GMATNIPPHN LGELVDVLCA LIHNPEATLQ 300
301 ELLEYMPAPD FPTGGIIMGN LGVLDAYRTG RGRVVVRGKA EVELLDPKTK RNAVIITEIP 360
361 YQTNKATLVQ KIAELVENKT LEGISDIRDE SDRNGMRVVI ELKRGGDPAL VLNNLYRHTA 420
421 LQSSFSCNMV GICDGEPKLM GLKELLQAFI DFRCSVVERR ARFKLSHAQQ RKHIIEGIVV 480
481 GLDNVDEVIE LITKASSHSS ATAALQSEYG LSEKQAEAIL EITLRRLTAL ERKKFTDESS 540
541 SLTEQITKLE QLLSTRTNIL KLIEQEAIEL KDRFSSPRRS MLEDSDSGDL EDIDVIPNEE 600
601 MLMAVSEKGY VKRMKADTFN LQHRGTIGKS VGKLRVDDAM SDFLVCHAHD HVLFFSDRGI 660
661 VYSTRAYKIP ECSRNAAGTP LVQILSMSEG ERVTSIVPVS EFAEDRYLLM LTVNGCIKKV 720
721 SLKLFSGIRS TGIIAIQLNS GDELKWVRCC SSDDLVAMAS QNGMVALSTC DGVRTLSRNT 780
781 KGVTAMRLKN EDKIASMDII PASLRKDMEE KSEDASLVKQ STGPWLLFVC ENGYGKRVPL 840
841 SSFRRSRLNR VGLSGYKFAE DDRLAAVFVV GYSLAEDGES DEQVVLVSQS GTVNRIKVRD 900
901 ISIQSRRARG VILMRLDHAG KIQSASLISA ADEEETEGTL SNEAVEAVSL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
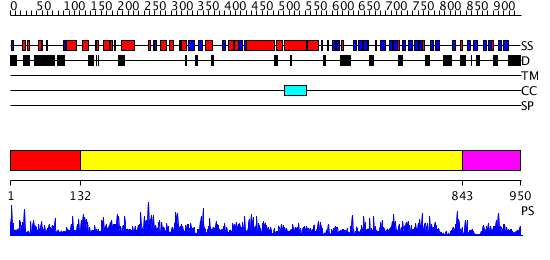
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..131] | 127.0 | No description for 2rgrA was found. | |
| 2 | View Details | [132..842] | 1000.0 | Structure of the full-length E. coli ParC subunit | |
| 3 | View Details | [843..950] | 98.522879 | A Superhelical Spiral in Escherichia coli DNA Gyrase A C-terminal Domain Imparts Unidirectional Supercoiling Bias |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.72 |
Source: Reynolds et al. (2008)