













| General Information: |
|
| Name(s) found: |
MBD4L_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 445 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
DNA repair
[IEA]
base-excision repair [IEA][ISS] |
| Molecular Function: |
catalytic activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVPPIIYKYK RRKDRRLGRD DDSSVMMTRR RPDSDFIEVS DENRSFALFK EDDEKNRDLG 60
61 LVDDGSTNLV LQCHDDGCSL EKDNSNSLDD LFSGFVYKGV RRRKRDDFGS ITTSNLVSPQ 120
121 IADDDDDSVS DSHIERQECS EFHVEVRRVS PYFQGSTVSQ QSKEGCDSDS VCSKEGCSKV 180
181 QAKVPRVSPY FQASTISQCD SDIVSSSQSG RNYRKGSSKR QVKVRRVSPY FQESTVSEQP 240
241 NQAPKGLRNY FKVVKVSRYF HADGIQVNES QKEKSRNVRK TPIVSPVLSL SQKTDDVYLR 300
301 KTPDNTWVPP RSPCNLLQED HWHDPWRVLV ICMLLNKTSG AQTRGVISDL FGLCTDAKTA 360
361 TEVKEEEIEN LIKPLGLQKK RTKMIQRLSL EYLQESWTHV TQLHGVGKYA ADAYAIFCNG 420
421 NWDRVKPNDH MLNYYWDYLR IRYKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
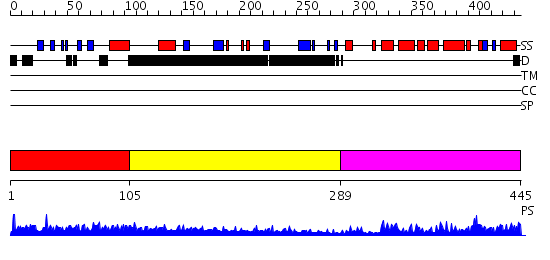
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..104] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [105..288] | 5.166992 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [289..445] | 28.154902 | Mismatch-specific thymine glycosylase domain of the methyl-GpG binding protein mbd4 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)