













| General Information: |
|
| Name(s) found: |
WRK33_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 519 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
response to cold
[IEP]
response to heat [IEP] camalexin biosynthetic process [IMP] response to salt stress [IMP] response to osmotic stress [IEP] regulation of transcription, DNA-dependent [ISS] defense response to fungus [IMP] response to chitin [IEP] defense response to bacterium [IMP] response to water deprivation [IEP] |
| Molecular Function: |
transcription factor activity
[IDA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAASFLTMDN SRTRQNMNGS ANWSQQSGRT STSSLEDLEI PKFRSFAPSS ISISPSLVSP 60
61 STCFSPSLFL DSPAFVSSSA NVLASPTTGA LITNVTNQKG INEGDKSNNN NFNLFDFSFH 120
121 TQSSGVSAPT TTTTTTTTTT TTNSSIFQSQ EQQKKNQSEQ WSQTETRPNN QAVSYNGREQ 180
181 RKGEDGYNWR KYGQKQVKGS ENPRSYYKCT FPNCPTKKKV ERSLEGQITE IVYKGSHNHP 240
241 KPQSTRRSSS SSSTFHSAVY NASLDHNRQA SSDQPNSNNS FHQSDSFGMQ QEDNTTSDSV 300
301 GDDEFEQGSS IVSRDEEDCG SEPEAKRWKG DNETNGGNGG GSKTVREPRI VVQTTSDIDI 360
361 LDDGYRWRKY GQKVVKGNPN PRSYYKCTTI GCPVRKHVER ASHDMRAVIT TYEGKHNHDV 420
421 PAARGSGYAT NRAPQDSSSV PIRPAAIAGH SNYTTSSQAP YTLQMLHNNN TNTGPFGYAM 480
481 NNNNNNSNLQ TQQNFVGGGF SRAKEEPNEE TSFFDSFMP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
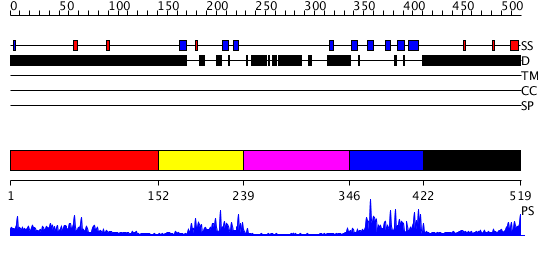
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..151] | 8.092992 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [152..238] | 5.1 | Solution Structure of the C-terminal WRKY Domain of AtWRKY4 | |
| 3 | View Details | [239..345] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [346..421] | 31.09691 | Solution Structure of the C-terminal WRKY Domain of AtWRKY4 | |
| 5 | View Details | [422..519] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)