













| General Information: |
|
| Name(s) found: |
DNJ13_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 538 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
response to very low light intensity stimulus
[IMP]
[NOT]response to high light intensity [IMP] vegetative to reproductive phase transition of meristem [IMP] protein folding [ISS] |
| Molecular Function: |
heat shock protein binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMGQEAAPTG PPNRELYALL NLSPEASDEE IRKAYRQWAQ VYHPDKIQSP QMKEVATENF 60
61 QRICEAYEIL SDETKRLIYD LYGMEGLNSG LELGPRLSKA DEIKEELERI KRRNEEAKKM 120
121 AHFQPTGSIL FNLSVPHFLV GDGIMRGMVM ASQVQSQLSK DDAIAIGGNL AANEKSGGGV 180
181 ATAILRRQIS PVSSIEFVAS TGLQSLIGVQ TTRQLTIHST ATINISKSLS DGSINLTNTW 240
241 TRQLSETSSG NIELALGMRS AITVGWKKRD ENVSAAGDFK IESGGLGASA RYTRKLSSKS 300
301 HGRIVGRIGS NALEIELGGG RQISEFSTVR MMYTVGLKGI FWKVELHRGS QKLIVPILLS 360
361 AHLAPVFATG AFIVPTSLYF LLKKFVVKPY LLKREKQKAL ENMEKTWGQV GEARARAEKA 420
421 QQLLQTVATR KKNRQVETDG LIVTKALYGD PKAIERRNEG VEGLDSGVID VTVPMNFLVS 480
481 DSGQLKLHEG VKKSGIMGFC DPCPGQPKQL YIAYTYHSQP FEVIVGDYEE LSIPQEGQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
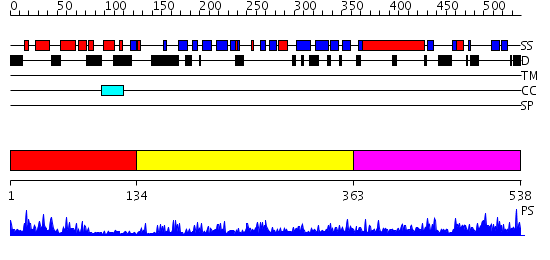
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..133] | 26.69897 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 2 | View Details | [134..362] | 35.39794 | The crystal structure of Hsp40 Ydj1 | |
| 3 | View Details | [363..538] | 2.237958 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|