













| General Information: |
|
| Name(s) found: |
INP54 /
YOL065C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 384 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum
[IDA |
| Biological Process: |
inositol lipid-mediated signaling
[TAS exocytosis [TAS phosphoinositide dephosphorylation [TAS |
| Molecular Function: |
phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNKTNWKVSV TTFNCGKEFP VENSKAIVKQ LLFPYDDGIS QLELQDLYVL GFQEVVPIWQ 60
61 GSFPAVNRDL IDRITTTAVN CLNEKVSATQ GDEQYSCLGV NSLGAITIIV LYNNNALKVK 120
121 DDILKRNGKC GWFGTHLKGG TLISFQMTRN GEENWERFSY ICAHLNANEG VNNRNQRIDD 180
181 YKRIMSEVCD SEVAKSDHFF FLGDLNFRVT STYDPTTNYS STTTLRRLLE NHEELNLLRK 240
241 GEDEPLCKGF QELKITFPPT YKFKLFEKET YNTKRIPSWC DRILYKSYAV PTFAQEGTYH 300
301 SVPRSNALLF SDHQPVNLTV RLPRSTGTPV PLSLHIEKYP LSWSSGLIGQ IGDAVIGYCG 360
361 WLVTKNVHYW ILGSLLLYLL LKIL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
CAP1 DFR1 INP54 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
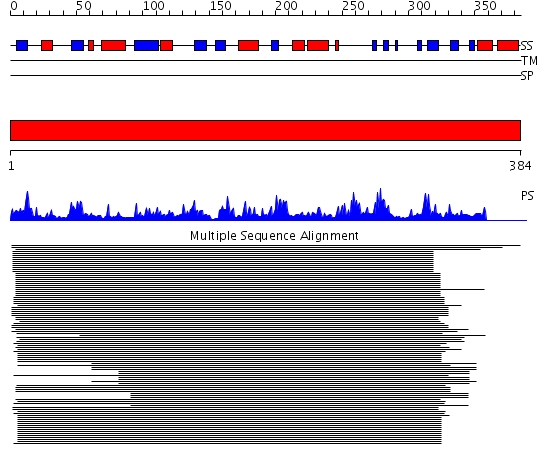
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..384] | 295.522879 | Synaptojanin, IPP5C domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.74 |
Source: Reynolds et al. (2008)