













| General Information: |
|
| Name(s) found: |
SPE2 /
YOL052C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 396 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
pantothenate biosynthetic process
[IMP spermidine biosynthetic process [IMP spermine biosynthetic process [IMP |
| Molecular Function: |
adenosylmethionine decarboxylase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTVTIKELTN HNYIDHELSA TLDSTDAFEG PEKLLEIWFF PHKKSITTEK TLRNIGMDRW 60
61 IEILKLVKCE VLSMKKTKEL DAFLLSESSL FVFDHKLTMK TCGTTTTLFC LEKLFQIVEQ 120
121 ELSWAFRTTQ GGKYKPFKVF YSRRCFLFPC KQAAIHQNWA DEVDYLNKFF DNGKSYSVGR 180
181 NDKSNHWNLY VTETDRSTPK GKEYIEDDDE TFEVLMTELD PECASKFVCG PEASTTALVE 240
241 PNEDKGHNLG YQMTKNTRLD EIYVNSAQDS DLSFHHDAFA FTPCGYSSNM ILAEKYYYTL 300
301 HVTPEKGWSY ASFESNIPVF DISQGKQDNL DVLLHILNVF QPREFSMTFF TKNYQNQSFQ 360
361 KLLSINESLP DYIKLDKIVY DLDDYHLFYM KLQKKI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
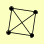
|
MRS1 RPL42B, RPL42A SEO1 SPE2 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |

Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..199] | 487.0 | S-adenosylmethionine decarboxylase | |
| 2 | View Details | [200..396] | 487.0 | S-adenosylmethionine decarboxylase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)