













| General Information: |
|
| Name(s) found: |
DOM34 /
YNL001W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 386 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
meiosis
[TAS translation [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKVISLKKDS FNKGGAVITL LPEDKEDLFT VYQIVDKDDE LIFKKKFTSK LDEAGKKKST 60
61 DLVKLKIKVI SEDFDMKDEY LKYKGVTVTD ESGASNVDIP VGKYLSFTLD YVYPFTIIKQ 120
121 NFNKFMQKLL NEACNIEYKS DTAAVVLQEG IAHVCLVTSS STILKQKIEY SMPKKKRTTD 180
181 VLKFDEKTEK FYKAIYSAMK KDLNFDKLKT IILCSPGFYA KILMDKIFQY AEEEHNKKIL 240
241 DNKGMFFIAH CSTGYLQGIN EVLKNPLYAS KLQDTKYSKE IMVMDEFLLH LNKDDDKAWY 300
301 GEKEVVKAAE YGAISYLLLT DKVLHSDNIA QREEYLKLMD SVESNGGKAL VLSTLHSLGE 360
361 ELDQLTGIAC ILKYPLPDLD EDDGEE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
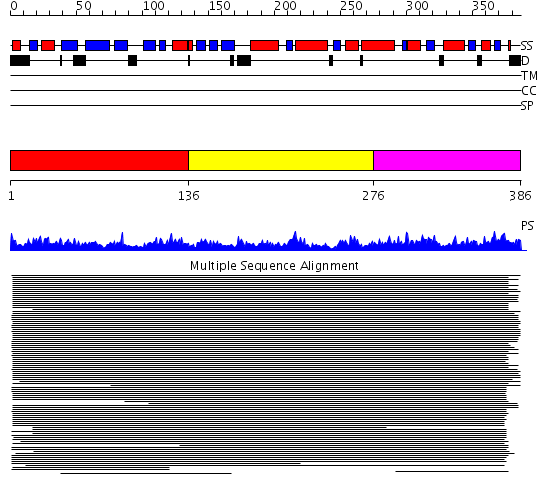
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..135] | 120.0 | Middle domain of eukaryotic peptide chain release factor subunit 1, ERF1; C-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1; N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1 | |
| 2 | View Details | [136..275] | 120.0 | Middle domain of eukaryotic peptide chain release factor subunit 1, ERF1; C-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1; N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1 | |
| 3 | View Details | [276..386] | 120.0 | Middle domain of eukaryotic peptide chain release factor subunit 1, ERF1; C-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1; N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)