













| General Information: |
|
| Name(s) found: |
VPS9 /
YML097C
[SGD]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 451 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
telomere maintenance
[IMP protein targeting to vacuole [IMP |
| Molecular Function: |
guanyl-nucleotide exchange factor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTDDEKREIL KEFDPFSQLE QANGNPDKDV KFKKDDPNRA AAEETNRDIS AQDKGDEEPF 60
61 YDFQIFIKQL QTPGADPLVK YTKSFLRNFL AQRLLWTVSE EIKLISDFKT FIYDKFTLYE 120
121 PFRSLDNSKM RNAKEGMEKL IMGKLYSRCF SPSLYEILQK PLDDEHMKDL TNDDTLLEKI 180
181 RHYRFISPIM LDIPDTMPNA RLNKFVHLAS KELGKINRFK SPRDKMVCVL NASKVIFGLL 240
241 KHTKLEQNGA DSFIPVLIYC ILKGQVRYLV SNVNYIERFR SPDFIRGEEE YYLSSLQAAL 300
301 NFIMNLTERS LTIEDHEDFE EAYQRNFKQL AEEKEEEEKK KQLEIPDELQ PNGTLLKPLD 360
361 EVTNIVISKF NELFSPIGEP TQEEALKSEQ SNKEEDVSSL IKKIEENERK DTLNTLQNMF 420
421 PDMDPSLIED VCIAKKSRIG PCVDALLSLS E |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2002) |
|
EXO84 SEC10 SEC3 SEC5 SEC6 SEC8 VPS9 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
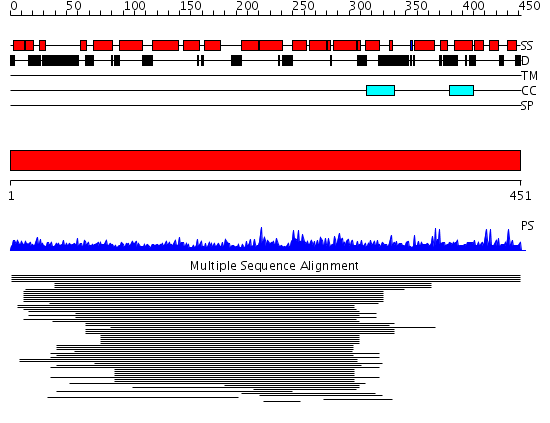
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..451] | 8.62 | (+)-bornyl diphosphate synthase | |
| 2 | View Details | [343..451] | 20.522879 | Vacuolar protein sorting-associated protein vps9 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)