













| General Information: |
|
| Name(s) found: |
SCW4 /
YGR279C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 386 amino acids |
Gene Ontology: |
|
| Cellular Component: |
fungal-type cell wall
[IDA |
| Biological Process: |
conjugation with cellular fusion
[IGI |
| Molecular Function: |
glucosidase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRLSNLIASA SLLSAATLAA PANHEHKDKR AVVTTTVQKQ TTIIVNGAAS TPVAALEENA 60
61 VVNSAPAAAT STTSSAASVA TAAASSSENN SQVSAAASPA SSSAATSTQS SSSSQASSSS 120
121 SSGEDVSSFA SGVRGITYTP YESSGACKSA SEVASDLAQL TDFPVIRLYG TDCNQVENVF 180
181 KAKASNQKVF LGIYYVDQIQ DGVNTIKSAV ESYGSWDDVT TVSIGNELVN GNQATPSQVG 240
241 QYIDSGRSAL KAAGYTGPVV SVDTFIAVIN NPELCDYSDY MAVNAHAYFD KNTVAQDSGK 300
301 WLLEQIQRVW TACDGKKNVV ITESGWPSKG ETYGVAVPSK ENQKDAVSAI TSSCGADTFL 360
361 FTAFNDYWKA DGAYGVEKYW GILSNE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
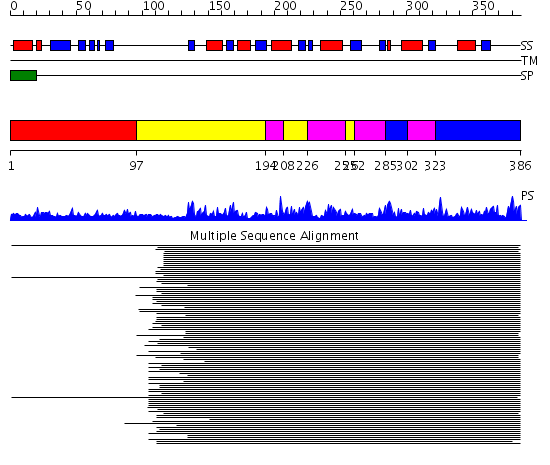
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..96] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [97..193] [208..225] [255..261] |
71.30103 | Plant beta-glucanases | |
| 3 | View Details | [194..207] [226..254] [262..284] [302..322] |
71.30103 | Plant beta-glucanases | |
| 4 | View Details | [285..301] [323..386] |
71.30103 | Plant beta-glucanases |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|