













| General Information: |
|
| Name(s) found: |
MRF1 /
YGL143C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 413 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
translational termination
[IMP translation [IMP |
| Molecular Function: |
translation release factor activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MWLSKFQFPS RSIFKGVFLG HKLPLLVRLT STTTNSKSNG SIPTQYTELS PLLVKQAEKY 60
61 EAELKDLDKD LSCGIHFDVN KQKHYAKLSA LTDTFIEYKE KLNELKSLQE MIVSDPSLRA 120
121 EAEQEYAELV PQYETTSSRL VNKLLPPHPF ADKPSLLELR PGVGGIEAMI FTQNLLDMYI 180
181 GYANYRKWKY RIISKNENES GSGIIDAILS IEEAGSYDRL RFEAGVHRVQ RIPSTETKGR 240
241 THTSTAAVVV LPQIGDESAK SIDAYERTFK PGEIRVDIMR ASGKGGQHVN TTDSAVRLTH 300
301 IPSGIVVSMQ DERSQHKNKA KAFTILRARL AEKERLEKEE KERKARKSQV SSTNRSDKIR 360
361 TYNFPQNRIT DHRCGFTLLD LPGVLSGERL DEVIEAMSKY DSTERAKELL ESN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
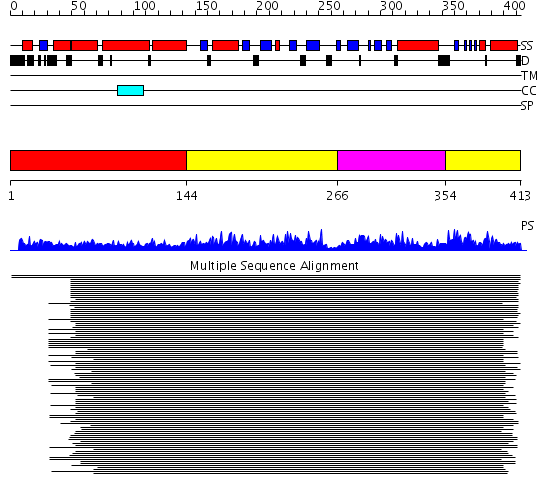
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..143] | 502.0 | Polypeptide chain release factor 2 (RF2) | |
| 2 | View Details | [144..265] [354..413] |
502.0 | Polypeptide chain release factor 2 (RF2) | |
| 3 | View Details | [266..353] | 502.0 | Polypeptide chain release factor 2 (RF2) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.69 |
Source: Reynolds et al. (2008)