













| General Information: |
|
| Name(s) found: |
PEP7 /
YDR323C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 515 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA external side of endosome membrane [IDA |
| Biological Process: |
vesicle fusion
[IMP vesicle docking during exocytosis [IMP Golgi to vacuole transport [IGI |
| Molecular Function: |
phosphatidylinositol-3-phosphate binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLENVSCPI CLRKFDNLQA LNAHLDVEHG FNDNEDSLGS NDSRLVNGKQ KKARSVDSSA 60
61 QKLKRSHWEK FKKGKSCCHT CGRTLNNNIG AINCRKCGKL YCRRHLPNMI KLNLSAQYDP 120
121 RNGKWYNCCH DCFVTKPGYN DYGEVIDLTP EFFKVRNIKR EDKNLRLLQL ENRFVRLVDG 180
181 LITLYNTYSR SIIHNLKMNS EMSKLERTVT PWRDDRSVLF CNICSEPFGL LLRKHHCRLC 240
241 GMVVCDDANR NCSNEISIGY LMSAASDLPF EYNIQKDDLL HIPISIRLCS HCIDMLFIGR 300
301 KFNKDVRMPL SGIFAKYDSM QNISKVIDSL LPIFEDSLNS LKVETAKDSE NTLDPKNLND 360
361 LARLRHKLLN SFNLYNTLTR QLLSVEPQSH LERQLQNSIK IASAAYINEK ILPLKSLPAI 420
421 LNPEGHKTNE DGQKAEPEVK KLSQLMIENL TIKEVKELRE ELMVLKEQSY LIESTIQDYK 480
481 KQRRLEEIVT LNKNLEELHS RIHTVQSKLG DHGFN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |

|
PEP3 PEP7 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
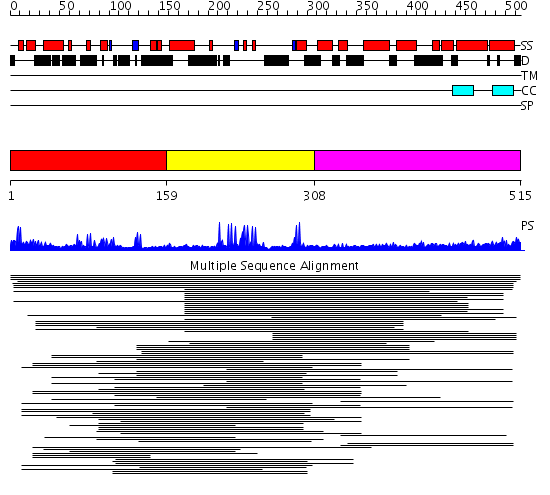
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..158] | 10.91 | Hrs | |
| 2 | View Details | [159..307] | 53.69897 | vps27p protein | |
| 3 | View Details | [308..515] | 12.085997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)