













| General Information: |
|
| Name(s) found: |
SWF1 /
YDR126W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 336 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear membrane-endoplasmic reticulum network
[IDA integral to membrane [ISS |
| Biological Process: |
protein amino acid palmitoylation
[IMP vacuole fusion, non-autophagic [IGI ascospore wall assembly [IMP |
| Molecular Function: |
protein-cysteine S-palmitoleyltransferase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSWNLLFVLL IGFVVLILLS PVFKSTWPFS TFYRNVFQPF LVDDQKYRWK LHLVPLFYTS 60
61 IYLYLVYTYH MRVESTIKNE LFLLERILIV PIIILPPVAL GILAMVSRAE DSKDHKSGST 120
121 EEYPYDYLLY YPAIKCSTCR IVKPARSKHC SICNRCVLVA DHHCIWINNC IGKGNYLQFY 180
181 LFLISNIFSM CYAFLRLWYI SLNSTSTLPR AVLTLTILCG CFTIICAIFT YLQLAIVKEG 240
241 MTTNEQDKWY TIQEYMREGK LVRSLDDDCP SWFFKCTEQK DDAAEPLQDQ HVTFYSTNAY 300
301 DHKHYNLTHY ITIKDASEIP NIYDKGTFLA NLTDLI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
|
ERF2 SHR5 SWF1 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
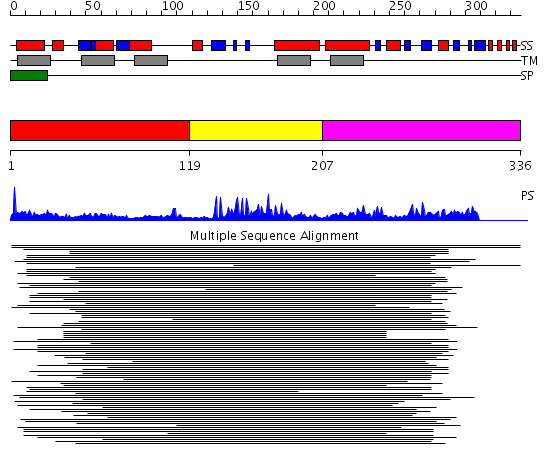
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..118] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [119..206] | 34.920819 | DHHC zinc finger domain No confident structure predictions are available. | |
| 3 | View Details | [207..336] | 1.132996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|