













| General Information: |
|
| Name(s) found: |
LYS20 /
YDL182W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 428 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mitochondrion [IDA |
| Biological Process: |
lysine biosynthetic process via aminoadipic acid
[TAS |
| Molecular Function: |
homocitrate synthase activity
[IEP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTAAKPNPYA AKPGDYLSNV NNFQLIDSTL REGEQFANAF FDTEKKIEIA RALDDFGVDY 60
61 IELTSPVASE QSRKDCEAIC KLGLKAKILT HIRCHMDDAK VAVETGVDGV DVVIGTSKFL 120
121 RQYSHGKDMN YIAKSAVEVI EFVKSKGIEI RFSSEDSFRS DLVDLLNIYK TVDKIGVNRV 180
181 GIADTVGCAN PRQVYELIRT LKSVVSCDIE CHFHNDTGCA IANAYTALEG GARLIDVSVL 240
241 GIGERNGITP LGGLMARMIV AAPDYVKSKY KLHKIRDIEN LVADAVEVNI PFNNPITGFC 300
301 AFTHKAGIHA KAILANPSTY EILDPHDFGM KRYIHFANRL TGWNAIKARV DQLNLNLTDD 360
361 QIKEVTAKIK KLGDVRSLNI DDVDSIIKNF HAEVSTPQVL SAKKNKKNDS DVPELATIPA 420
421 AKRTKPSA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
LYS20 LYS21 RPL18A, RPL18B RPL2A, RPL2B RPS1A RPS3 |
| View Details | Krogan NJ, et al. (2006) |

|
GLO3 ISA2 KEX1 LYS20 RPL37B YBL086C |
| View Details | Qiu et al. (2008) |

|
LYS20 LYS21 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
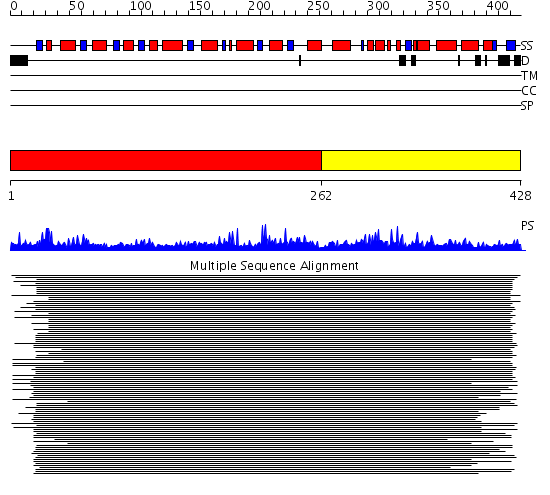
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..261] | 11.45 | KDPG aldolase | |
| 2 | View Details | [262..428] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)