













| General Information: |
|
| Name(s) found: |
FBpp0309007
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 1945 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mRNA cleavage factor complex
[ISS]
|
| Biological Process: |
mRNA cleavage
[ISS]
|
| Molecular Function: |
RNA binding
[IDA]
negative transcription elongation factor activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEQLFQNYRD DERRIGEEYL SSLQDLNCNS KPLINMLTML AEENINYAHI IVKVVEYYIS 60
61 QVAPEFKLPI LYLIDSIVKN VKSSYVQLFG QCIVNIFLHA FESVQHSQSQ VLEKVRERMY 120
121 ALRQTWNEVF PPSKMYALDV KVKRLDNNWP ITAKQPTNKI HVNPAIHVNP DFLKPGLVPG 180
181 MPGNPTITSD MEEILQAKTR ELLELKKRKL ELELEQTKKH LEEQERQLTQ TTDAMVGAPI 240
241 IMPAPTPAAI CPPITDPGIA VHNQRAPGVM GNVGPVMHNM PVAPQLFANK PKVHPVNPAL 300
301 LNSVRQRDPR LARQMHAASS HPAAPARNDP RLEAKSSSSQ KSSRSRSKSP VRNSSSRSGK 360
361 SGSSSHSSSS SRKRSESKSS TTSSSSSSSD VRHKGVTASS SNQPTVKRSG KITSKDSDRY 420
421 VRNGSPLGSA KRKSSSPSSS PSKSKRSTSH KSSSSRGKTS SLRSRSRSPV FMDVDLRTGV 480
481 RSKSPESRAV APSSLPLAGA SLPASAKAPK DLEKRHPPAP PSPPIIDVSS MDIDLRRPKT 540
541 PPPPTFDSVD SLDQKDTTSG HISSTIIQAS GASNASPSGA STSKLPAKVS FKIQKQFNKL 600
601 EKSTANLSTV SMLNSNPTTS GTPAASLVRQ SSPSPSSSSI SSQGNVSDAL QLSINKLLQV 660
661 QGGAGEKRPA DALPPHIDGE EQPPQQKRSK SAKLDALFGS EDVDLRREIL VVKPGVIVVE 720
721 DDSMDECDVD KPTKKSGSPP KKATLEELRA KLANSARIQN KQGKSDKSRD QAVSQRLKQL 780
781 AELKVNDDSQ EAHDEKVRTI LSQAQEMYEN NGMNQEQYKD LVQKVVAINE NSKIKESRRR 840
841 DNDLERNAAR DAVLRKRIPK LKGNENHSAG SPHTDGSPRY DDQPAAAPEK PSNKRDKTKR 900
901 EMKRRKPTKW GEQVDAGAAQ RTAWQLANVN NNNNNNNNKR GGIQLPVQPQ AGFRGMPWQQ 960
961 PPAMVLGQSA VPPPPQPPSM ISMPPVPPVT MTKAINSLDN PMADVVRSIT IDGGSKEIRF1020
1021 YNQVAIIFMD GDQPHEIGFQ PGQRVIFIDH NEPLPLCFND DYKPFHLDGQ LHRIRFGFPS1080
1081 RELYIDEHWY EIYFGGPAVS VPIGNKLHII KAEGPPPNVD IGRVRRDLVV GKINMIADAH1140
1141 TIVPLFLDAR QQTFQLGAEQ HSLQFVDSFQ YALLDGQLQK MEYGGLPKGM MLNGGRSCFI1200
1201 RFGTLPKGVI AGKTHVADMV YIKTDAPAEP PKPPPIIVKP PPVVEQPKPA PVAVAPISLP1260
1261 AAAAALESLN INDLFQKLVS SGIIGGATAA PTLPAADSSA AKEPTASATE PSTASATVAP1320
1321 ATLPAGPPAE PIKRIDLRKP ESIKTRQAAV VAVLYLGMQC SSCGVRFPPE QTIKYSQHLD1380
1381 WHFRQNRRER DSTRKATSRK WYYDLNDWRQ YEEIEDVEER EKNFLEAQGQ PGGVEALDEL1440
1441 SQQRSLDSPV PTCAAGRDDV DHCCDMCHEK FEQFYNEELE EWHLRSAIRV EDKIYHPLCY1500
1501 EDYKASLNPP TEVKSDQDVD MNNTDDNAMD TLIKVEDDED EGTPNSSQPI FEDDDDDVIV1560
1561 LPNEEPSVTE IVDDDDEDEY VPGNVTRADM GNESQDKTES NSETKEPKKD QVELGEQQQN1620
1621 ESANESDVEI QEPNIPFTDL DTYVEKEMDE ATRAALLNVK IKEEPKDEYE EDEDDGFEDV1680
1681 GTVVSLLPLP EDEISIHSSE TQTHTIGSSA SPATIERPAS VTSLSLPANE ADELEQGDAN1740
1741 TDTDAELNGE KQEATHNLSA VGPALPLASI VNKIKINITK NTSSNSHNSA SNTTTTDSQV1800
1801 SAISVIGGSG AAGGASGAGD PVQQVNAIQT ISTIPVLCGG NTFVPKIATS TPSNVISSIS1860
1861 VIGSTYGASS SSNGSRSTTS TASSAPAASL LAETGSLKSP TPPLATVDPD PEPVVEQKPA1920
1921 LRNATLKRTK KVQNGIETSG LCSIM |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
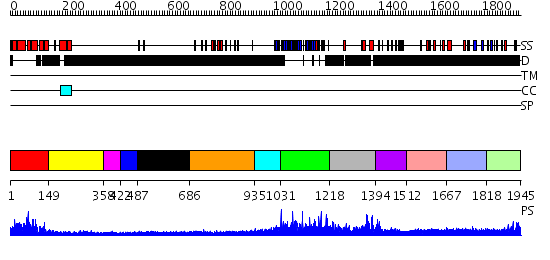
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..148] | 32.522879 | The RNA polymerase II CTD in mRNA processing: beta-turn recognition and beta-spiral model | |
| 2 | View Details | [149..357] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [358..421] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [422..486] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [487..685] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [686..934] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [935..1030] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1031..1217] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1218..1393] | 1.016991 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1394..1511] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1512..1666] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1667..1817] | 2.080996 | View MSA. No confident structure predictions are available. | |
| 13 | View Details | [1818..1945] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||
| 10 | No functions predicted. | |||||||||||||||||||||
| 11 | No functions predicted. | |||||||||||||||||||||
| 12 | No functions predicted. | |||||||||||||||||||||
| 13 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)