













| General Information: |
|
| Name(s) found: |
gi|9714268
[NCBI NR]
gi|24234753 [NCBI NR] gi|119604537 [NCBI NR] |
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 702 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRPMRIFVND DRHVMAKHSS VYPTQEELEA VQNMVSHTER ALKAVSDWID EQEKGSSEQA 60
61 ESDNMDVPPE DDSKEGAGEQ KTEHMTRTLR GVMRVGLVAK GLLLKGDLDL ELVLLCKEKP 120
121 TTALLDKVAD NLAIQLAAVT EDKYEILQSV DDAAIVIKNT KEPPLSLTIH LTSPVVREEM 180
181 EKVLAGETLS VNDPPDVLDR QKCLAALASL RHAKWFQARA NGLKSCVIVI RVLRDLCTRV 240
241 PTWGPLRGWP LELLCEKSIG TANRPMGAGE ALRRVLECLA SGIVMPDGSG IYDPCEKEAT 300
301 DAIGHLDRQQ REDITQSAQH ALRLAAFGQL HKVLGMDPLP SKMPKKPKNE NPVDYTVQIP 360
361 PSTTYAITPM KRPMEEDGEE KSPSKKKKKI QKKEEKAEPP QAMNALMRLN QLKPGLQYKL 420
421 VSQTGPVHAP IFTMSVEVDG NSFEASGPSK KTAKLHVAVK VLQDMGLPTG AEGRDSSKGE 480
481 DSAEETEAKP AVVAPAPVVE AVSTPSAAFP SDATAEQGPI LTKHGKNPVM ELNEKRRGLK 540
541 YELISETGGS HDKRFVMEVE VDGQKFQGAG SNKKVAKAYA ALAALEKLFP DTPLALDANK 600
601 KKRAPVPVRG GPKFAAKPHN PGFGMGGPMH NEVPPPPNLR GRGRGGSIRG RGRGRGFGGA 660
661 NHGGYMNAGA GYGSYGYGGN SATAGYSDFF TDCYGYHDFG SS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
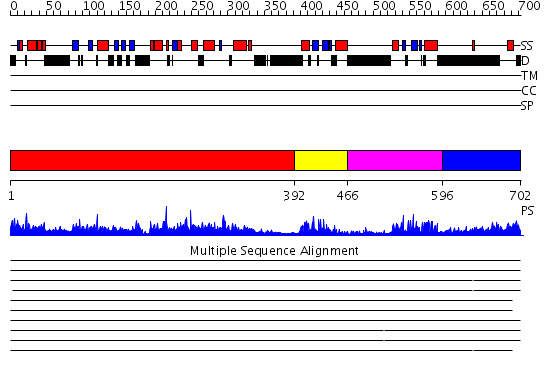
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..391] | 2.94 | Crystal structure of the 2'-specific and double-stranded RNA-activated interferon-induced antiviral protein 2'-5'-oligoadenylate synthetase | |
| 2 | View Details | [392..465] | 14.30103 | No description for 2dmyA was found. | |
| 3 | View Details | [466..595] | 4.06 | dsRNA-dependent protein kinase pkr | |
| 4 | View Details | [596..702] | 14.0 | Crystal structure of the catalytic domain of an adenosine deaminase that acts on RNA (hADAR2) bound to inositol hexakisphosphate (IHP) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)