













| General Information: |
|
| Name(s) found: |
PUTA_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 1320 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGTTTMGVKL DDATRERIKS AATRIDRTPH WLIKQAIFSY LEQLENSDTL PELPALLSGA 60
61 ANESDEAPTP AEEPHQPFLD FAEQILPQSV SRAAITAAYR RPETEAVSML LEQARLPQPV 120
121 AEQAHKLAYQ LADKLRNQKN ASGRAGMVQG LLQEFSLSSQ EGVALMCLAE ALLRIPDKAT 180
181 RDALIRDKIS NGNWQSHIGR SPSLFVNAAT WGLLFTGKLV STHNEASLSR SLNRIIGKSG 240
241 EPLIRKGVDM AMRLMGEQFV TGETIAEALA NARKLEEKGF RYSYDMLGEA ALTAADAQAY 300
301 MVSYQQAIHA IGKASNGRGI YEGPGISIKL SALHPRYSRA QYDRVMEELY PRLKSLTLLA 360
361 RQYDIGINID AEESDRLEIS LDLLEKLCFE PELAGWNGIG FVIQAYQKRC PLVIDYLIDL 420
421 ATRSRRRLMI RLVKGAYWDS EIKRAQMDGL EGYPVYTRKV YTDVSYLACA KKLLAVPNLI 480
481 YPQFATHNAH TLAAIYQLAG QNYYPGQYEF QCLHGMGEPL YEQVTGKVAD GKLNRPCRIY 540
541 APVGTHETLL AYLVRRLLEN GANTSFVNRI ADTSLPLDEL VADPVTAVEK LAQQEGQTGL 600
601 PHPKIPLPRD LYGHGRDNSA GLDLANEHRL ASLSSALLNS ALQKWQALPM LEQPVAAGEM 660
661 SPVINPAEPK DIVGYVREAT PREVEQALES AVNNAPIWFA TPPAERAAIL HRAAVLMESQ 720
721 MQQLIGILVR EAGKTFSNAI AEVREAVDFL HYYAGQVRDD FANETHRPLG PVVCISPWNF 780
781 PLAIFTGQIA AALAAGNSVL AKPAEQTPLI AAQGIAILLE AGVPPGVVQL LPGRGETVGA 840
841 QLTGDDRVRG VMFTGSTEVA TLLQRNIASR LDAQGRPIPL IAETGGMNAM IVDSSALTEQ 900
901 VVVDVLASAF DSAGQRCSAL RVLCLQDEIA DHTLKMLRGA MAECRMGNPG RLTTDIGPVI 960
961 DSEAKANIER HIQTMRSKGR PVFQAVRENS EDAREWQSGT FVAPTLIELD DFAELQKEVF1020
1021 GPVLHVVRYN RNQLPELIEQ INASGYGLTL GVHTRIDETI AQVTGSAHVG NLYVNRNMVG1080
1081 AVVGVQPFGG EGLSGTGPKA GGPLYLYRLL ANRPESALAV TLARQDAKYP VDAQLKAALT1140
1141 QPLNALREWA ANRPELQALC TQYGELAQAG TQRLLPGPTG ERNTWTLLPR ERVLCIADDE1200
1201 QDALTQLAAV LAVGSQVLWP DDALHRQLVK ALPSAVSERI QLAKAENITA QPFDAVIFHG1260
1261 DSDQLRALCE AVAARDGTIV SVQGFARGES NILLERLYIE RSLSVNTAAA GGNASLMTIG1320
1321 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
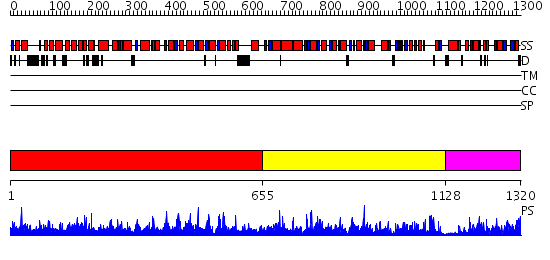
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..654] | 1000.0 | N-terminal domain of bifunctional PutA protein; Proline dehydrohenase domain of bifunctional PutA protein | |
| 2 | View Details | [655..1127] | 157.0 | Aldehyde reductase (dehydrogenase), ALDH | |
| 3 | View Details | [1128..1320] | 4.29 | 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | |||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)