













| General Information: |
|
| Name(s) found: |
NPD_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 279 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSRRGHRLS RFRKNKRRLR ERLRQRIFFR DKVVPEAMEK PRVLVLTGAG ISAESGIRTF 60
61 RAADGLWEEH RVEDVATPEG FDRDPELVQA FYNARRRQLQ QPEIQPNAAH LALAKLQDAL 120
121 GDRFLLVTQN IDNLHERAGN TNVIHMHGEL LKVRCSQSGQ VLDWTGDVTP EDKCHCCQFP 180
181 APLRPHVVWF GEMPLGMDEI YMALSMADIF IAIGTSGHVY PAAGFVHEAK LHGAHTVELN 240
241 LEPSQVGNEF AEKYYGPASQ VVPEFVEKLL KGLKAGSIA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
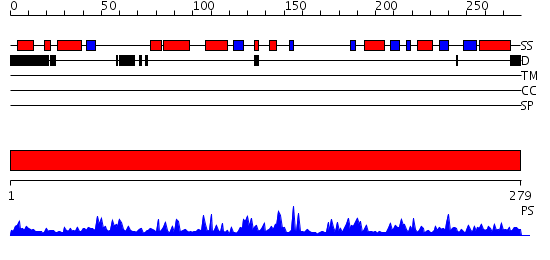
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..279] | 71.0 | Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Eschericia coli. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|