













| General Information: |
|
| Name(s) found: |
HSCA_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 616 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALLQISEPG LSAAPHQRRL AAGIDLGTTN SLVATVRSGQ AETLADHEGR HLLPSVVHYQ 60
61 QQGHSVGYDA RTNAALDTAN TISSVKRLMG RSLADIQQRY PHLPYQFQAS ENGLPMIETA 120
121 AGLLNPVRVS ADILKALAAR ATEALAGELD GVVITVPAYF DDAQRQGTKD AARLAGLHVL 180
181 RLLNEPTAAA IAYGLDSGQE GVIAVYDLGG GTFDISILRL SRGVFEVLAT GGDSALGGDD 240
241 FDHLLADYIR EQAGIPDRSD NRVQRELLDA AIAAKIALSD ADSVTVNVAG WQGEISREQF 300
301 NELIAPLVKR TLLACRRALK DAGVEADEVL EVVMVGGSTR VPLVRERVGE FFGRPPLTSI 360
361 DPDKVVAIGA AIQADILVGN KPDSEMLLLD VIPLSLGLET MGGLVEKVIP RNTTIPVARA 420
421 QDFTTFKDGQ TAMSIHVMQG ERELVQDCRS LARFALRGIP ALPAGGAHIR VTFQVDADGL 480
481 LSVTAMEKST GVEASIQVKP SYGLTDSEIA SMIKDSMSYA EQDVKARMLA EQKVEAARVL 540
541 ESLHGALAAD AALLSAAERQ VIDDAAAHLS EVAQGDDVDA IEQAIKNVDK QTQDFAARRM 600
601 DQSVRRALKG HSVDEV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
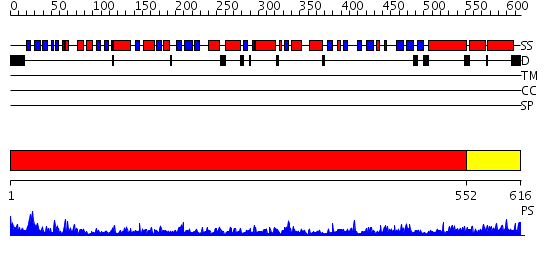
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..551] | 169.0 | crystal structure of bovine hsc70(aa1-554)E213A/D214A mutant | |
| 2 | View Details | [552..616] | 45.221849 | HscA substrate binding domain complexed with the IscU recognition peptide ELPPVKIHC |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)