













| General Information: |
|
| Name(s) found: |
BIOF_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 384 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSWQEKINAA LDARRAADAL RRRYPVAQGA GRWLVADDRQ YLNFSSNDYL GLSHHPQIIR 60
61 AWQQGAEQFG IGSGGSGHVS GYSVVHQALE EELAEWLGYS RALLFISGFA ANQAVIAAMM 120
121 AKEDRIAADR LSHASLLEAA SLSPSQLRRF AHNDVTHLAR LLASPCPGQQ MVVTEGVFSM 180
181 DGDSAPLAEI QQVTQQHNGW LMVDDAHGTG VIGEQGRGSC WLQKVKPELL VVTFGKGFGV 240
241 SGAAVLCSST VADYLLQFAR HLIYSTSMPP AQAQALRASL AVIRSDEGDA RREKLAALIT 300
301 RFRAGVQDLP FTLADSCSAI QPLIVGDNSR ALQLAEKLRQ QGCWVTAIRP PTVPAGTARL 360
361 RLTLTAAHEM QDIDRLLEVL HGNG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
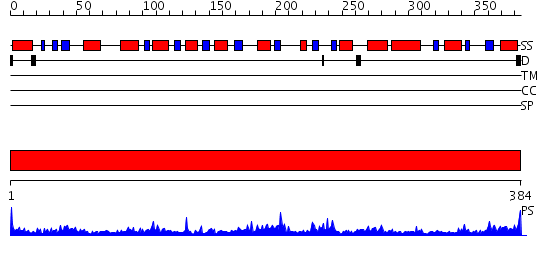
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..384] | 84.522879 | PLP-dependent acyl-CoA synthase (8-amino-7-oxonanoate synthase, AONS) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.71 |
Source: Reynolds et al. (2008)