













| General Information: |
|
| Name(s) found: |
BETA_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 556 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQFDYIIIGA GSAGNVLATR LTEDPNTSVL LLEAGGPDYR FDFRTQMPAA LAFPLQGKRY 60
61 NWAYETEPEP FMNNRRMECG RGKGLGGSSL INGMCYIRGN ALDLDNWAQE PGLENWSYLD 120
121 CLPYYRKAET RDMGENDYHG GDGPVSVTTS KPGVNPLFEA MIEAGVQAGY PRTDDLNGYQ 180
181 QEGFGPMDRT VTPQGRRAST ARGYLDQAKS RPNLTIRTHA MTDHIIFDGK RAVGVEWLEG 240
241 DSTIPTRATA NKEVLLCAGA IASPQILQRS GVGNAELLAE FDIPLVHELP GVGENLQDHL 300
301 EMYLQYECKE PVSLYPALQW WNQPKIGAEW LFGGTGVGAS NHFEAGGFIR SREEFAWPNI 360
361 QYHFLPVAIN YNGSNAVKEH GFQCHVGSMR SPSRGHVRIK SRDPHQHPAI LFNYMSHEQD 420
421 WQEFRDAIRI TREIMHQPAL DQYRGREISP GVECQTDEQL DEFVRNHAET AFHPCGTCKM 480
481 GYDEMSVVDG EGRVHGLEGL RVVDASIMPQ IITGNLNATT IMIGEKIADM IRGQEALPRS 540
541 TAGYFVANGM PVRAKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
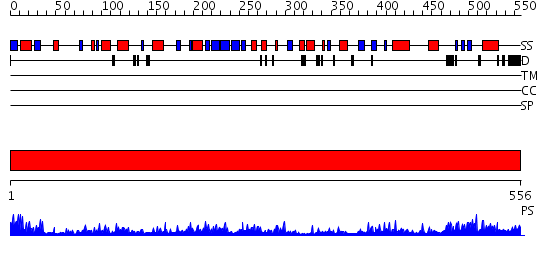
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..556] | 84.0 | Glucose oxidase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)