













| General Information: |
|
| Name(s) found: |
ATOC_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 461 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTAINRILIV DDEDNVRRML STAFALQGFE THCANNGRTA LHLFADIHPD VVLMDIRMPE 60
61 MDGIKALKEM RSHETRTPVI LMTAYAEVET AVEALRCGAF DYVIKPFDLD ELNLIVQRAL 120
121 QLQSMKKEIR HLHQALSTSW QWGHILTNSP AMMDICKDTA KIALSQASVL ISGESGTGKE 180
181 LIARAIHYNS RRAKGPFIKV NCAALPESLL ESELFGHEKG AFTGAQTLRQ GLFERANEGT 240
241 LLLDEIGEMP LVLQAKLLRI LQEREFERIG GHQTIKVDIR IIAATNRDLQ AMVKEGTFRE 300
301 DLFYRLNVIH LILPPLRDRR EDISLLANHF LQKFSSENQR DIIDIDPMAM SLLTAWSWPG 360
361 NIRELSNVIE RAVVMNSGPI IFSEDLPPQI RQPVCNAGEV KTAPVGERNL KEEIKRVEKR 420
421 IIMEVLEQQE GNRTRTALML GISRRALMYK LQEYGIDPAD V |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
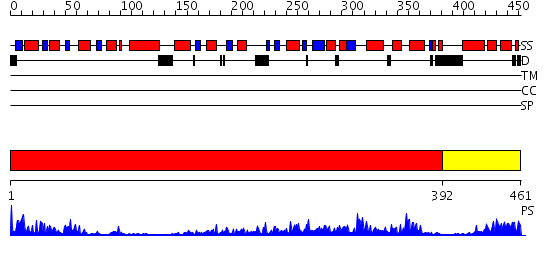
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..391] | 88.69897 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 2 | View Details | [392..461] | 84.154902 | Crystal structure of a sigma54-activator suggests the mechanism for the conformational switch necessary for sigma54 binding |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)