













| General Information: |
|
| Name(s) found: |
PAO3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 488 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisome
[IDA]
|
| Biological Process: |
polyamine catabolic process
[IDA]
|
| Molecular Function: |
polyamine oxidase activity
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MESGGKTNRQ LRKAICVSTD EKMKKKRSPS VIVIGGGMAG ISAARTLQDA SFQVVVLESR 60
61 DRIGGRVHTD YSFGFPVDLG ASWLHGVCKE NPLAAVIGRL GLPLYRTSGD NSVLYDHDLE 120
121 SYALFDKAGN QVSQELVTKV GENFEHILEE ICKVRDEQDE DMSIAQAFSI VFKRNPELRL 180
181 EGLAHNVLQW YLCRMEGWFA ADAETISAKC WDQEELLPGG HGLMVRGYRP VINTLSKGLD 240
241 IRLSHRITKI SRRYSGVKVT TEKGDTFVAD AAVIALPLGV LKSGMITFEP KLPQWKQEAI 300
301 NDLGVGIENK IILNFDNVFW PNVEFLGVVA ETSYGCSYFL NLHKATSHPV LVYMPAGQLA 360
361 RDIEKKSDEA AANFAFSQLQ KILPDASSPI NYLVSRWGSD INSLGSYSYD IVNKPHDLYE 420
421 RLRVPLDNLF FAGEATSSSY PGSVHGAYST GVLAAEDCRM RVLERYGELE HEMEEEAPAS 480
481 VPLLISRM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
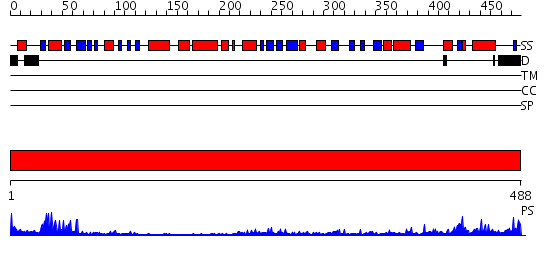
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..488] | 53.0 | Polyamine oxidase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.51 |
Source: Reynolds et al. (2008)