













| General Information: |
|
| Name(s) found: |
GRS17_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 488 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
cell redox homeostasis
[IEA]
|
| Molecular Function: |
protein disulfide oxidoreductase activity
[IEA]
electron carrier activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGTVKDIVS KAELDNLRQS GAPVVLHFWA SWCDASKQMD QVFSHLATDF PRAHFFRVEA 60
61 EEHPEISEAY SVAAVPYFVF FKDGKTVDTL EGADPSSLAN KVGKVAGSST SAEPAAPASL 120
121 GLAAGPTILE TVKENAKASL QDRAQPVSTA DALKSRLEKL TNSHPVMLFM KGIPEEPRCG 180
181 FSRKVVDILK EVNVDFGSFD ILSDNEVREG LKKFSNWPTF PQLYCNGELL GGADIAIAMH 240
241 ESGELKDAFK DLGITTVGSK ESQDEAGKGG GVSSGNTGLS ETLRARLEGL VNSKPVMLFM 300
301 KGRPEEPKCG FSGKVVEILN QEKIEFGSFD ILLDDEVRQG LKVYSNWSSY PQLYVKGELM 360
361 GGSDIVLEMQ KSGELKKVLT EKGITGEQSL EDRLKALINS SEVMLFMKGS PDEPKCGFSS 420
421 KVVKALRGEN VSFGSFDILT DEEVRQGIKN FSNWPTFPQL YYKGELIGGC DIIMELSESG 480
481 DLKATLSE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
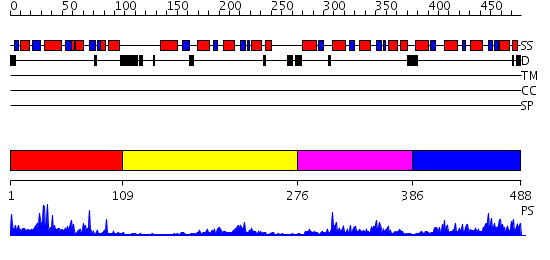
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..108] | 30.0 | No description for 2h71A was found. | |
| 2 | View Details | [109..275] | 26.070581 | No description for PF00462.15 was found. No confident structure predictions are available. | |
| 3 | View Details | [276..385] | 5.51 | Solution Structure of the PICOT homology 2 domain of the mouse PKC-interacting cousin of thioredoxin protein | |
| 4 | View Details | [386..488] | 31.0 | Solution structure of Grx4, a monothiol glutaredoxin from E. coli. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)