













| General Information: |
|
| Name(s) found: |
RH50_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 781 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA] ATP-dependent helicase activity [IEA][ISS] helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLARAPPPYF NFPARNNTIC NRNEIVRLFR NGGGVVARGA GFTRRPLETS SSYDDSTDDG 60
61 FVIISAADKE NEFAPPPSSD LLSSIPSESA RRNGSRSRGL TASFGRLKAQ KVKALVGKVT 120
121 QKKQHMSHNE EEDEDDASDE NYSADEGFGS SSILDLMRKK LAMKAIPRSG KSAERNEVKR 180
181 ASKVRESRES RRDLDRLEGD DEDVDEVSNP DRFTDNQRAG SRSSYSKGGY AANSRGKGDR 240
241 LSVARDLDSF EGHGRAIDEV SNPRKFNDNE RAESRSSYSR DSSANSRGRE DRRFVAKELD 300
301 TFQGRDKAYD EVYNPRRFTD NERGLRGGSH SKGSDTNSRG WGDRRSVVYT RDMDDWRERN 360
361 KTKDTRETGF FSRKTFAEIG CSEDMMKALK EQNFDRPAHI QAMAFSPVID GKSCIIADQS 420
421 GSGKTLAYLV PVIQRLREEE LQGHSKSSPG CPRVIVLVPT AELASQVLAN CRSISKSGVP 480
481 FRSMVVTGGF RQRTQLENLE QGVDVLIATP GRFTYLMNEG ILGLSNLRCA ILDEVDILFG 540
541 DDEFEAALQN LINSSPVTAQ YLFVTATLPL EIYNKLVEVF PDCEVVMGPR VHRVSNALEE 600
601 FLVDCSGDDN AEKTPETAFQ NKKTALLQIM EENPVSKTII FCNKIETCRK VENIFKRVDR 660
661 KERQLHVLPF HAALSQESRL TNMQEFTSSQ PEENSLFLVC TDRASRGIDF SGVDHVVLFD 720
721 FPRDPSEYVR RVGRTARGAR GKGKAFIFVV GKQVGLARRI IERNEKGHPV HDVPNAYEFT 780
781 T |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
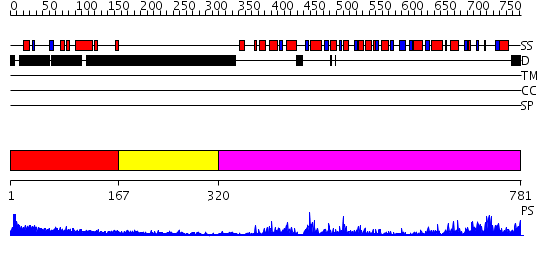
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..166] | 5.175997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [167..319] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [320..781] | 91.522879 | No description for 2db3A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)