













| General Information: |
|
| Name(s) found: |
G6PD5_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 516 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
cytosol [ISS] |
| Biological Process: |
pentose-phosphate shunt, oxidative branch
[IDA]
response to cadmium ion [IEP] glucose metabolic process [IDA][ISS] |
| Molecular Function: |
glucose-6-phosphate dehydrogenase activity
[IDA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSGQWHMEK RSTLKNDSFV KEYNPVTETG SLSIIVLGAS GDLAKKKTFP ALFNLFHQGF 60
61 LNPDEVHIFG YARSKITDEE LRDKIRGYLV DEKNASKKTE ALSKFLKLIK YVSGPYDSEE 120
121 GFKRLDKAIL EHEISKKTAE GSSRRLFYLA LPPSVYPPVS KMIKAWCTNK SDLGGWTRIV 180
181 VEKPFGKDLE SAEQLSSQIG ALFEEPQIYR IDHYLGKELV QNMLVLRFAN RLFLPLWNRD 240
241 NIANVQIVFR EDFGTEGRGG YFDEYGIIRD IIQNHLLQVL CLVAMEKPIS LKPEHIRDEK 300
301 VKVLQSVIPI KDEEVVLGQY EGYRDDPTVP NDSNTPTFAT TILRINNERW EGVPFILKAG 360
361 KAMSSKKADI RIQFKDVPGD IFKCQNQGRN EFVIRLQPSE AMYMKLTVKQ PGLEMQTVQS 420
421 ELDLSYKQRY QDVSIPEAYE RLILDTIRGD QQHFVRRDEL KAAWEIFTPL LHRIDKGEVK 480
481 SVPYKQGSRG PAEADQLLKK AGYMQTHGYI WIPPTL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
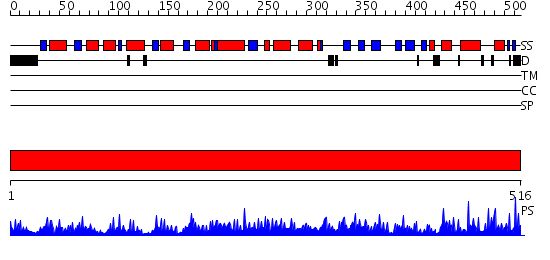
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..516] | 1000.0 | Glucose 6-phosphate dehydrogenase, N-terminal domain; Glucose 6-phosphate dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)