













| General Information: |
|
| Name(s) found: |
nhr-34 /
CE35564
[WormBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 613 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
regulation of transcription, DNA-dependent
[IEA |
| Molecular Function: |
DNA binding
[IEA transcription factor activity [IEA zinc ion binding [IEA steroid hormone receptor activity [IEA sequence-specific DNA binding [IEA steroid binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMDDLHQQLH QNYNPNGSPT TSTTHRRQNS SSGLEPARKR PKLSPPVLTA MPLDLTDAVS 60
61 PNESQACTSS LKGVVTAAGA AQMSVADRMT SSSVAPPLCI VPRSVEFINP KTEDMSMDIP 120
121 MGNVQSEFTG LKCRVCGDSR AGRHYGTIAC NGCKGFFRRS IWEQRDYVCR FGGKCLVVQE 180
181 YRNRCRACRL RKCFTVGMDA RAVQSERDKH KKNPKDSNNE GSTSPQYPTA STPISIPSTS 240
241 TSQTPTSSVN SYNFQNIPGI VSRSFSENLI MRDNSVPVME TSQSAALSHV PLVRYLIDLE 300
301 KATDNLIDEN CDFMSMEFDQ LCRVDVTIEA AFRQPGVVAK RTPPRWLALE RLTTLEDVHI 360
361 AWCRSFVLCI DYAKIMKDYQ ELSPTDQFTL LRNRVISVNW LCHTYKTFKA GCDGVALVNG 420
421 SWYPRDKELQ KQLDPGCNHY FRILSEHLME DLVIPMREMD MDEGEFVILK ALILFRAHRR 480
481 LSEEGRAHIK RVRDKYIEAL YQHVQHQHRH FSSVQTSMRI SKILLLLPSI EHLSQQEDDN 540
541 VQFLALFNLA NLNGLPYELH SSIKQHIPNG DDSDDTQVNE VTSNNDGPRS SESSHTPQSV 600
601 STSQFLEFKP SLH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
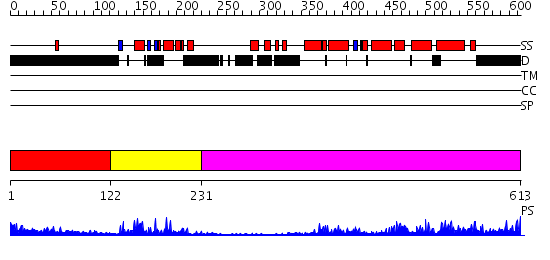
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | 1.258994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [122..230] | 26.69897 | Human Liver Receptor Homologue DNA-Binding Domain (hLRH-1 DBD) in Complex with dsDNA from the hCYP7A1 Promoter | |
| 3 | View Details | [231..613] | 62.39794 | LIGAND-BINDING DOMAIN OF THE HUMAN NUCLEAR RECEPTOR RXR-ALPHA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)