













| General Information: |
|
| Name(s) found: |
tbx-7 /
CE29639
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 327 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA transcription factor complex [ISS |
| Biological Process: |
regulation of transcription, DNA-dependent
[IEA regulation of transcription from RNA polymerase II promoter [ISS |
| Molecular Function: |
transcription factor activity
[IEA RNA polymerase II transcription factor activity, enhancer binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEGYGRSLF SEKHFPHDRQ LLPPFTTTMS LGSFFSFPAS SAFFDRKKDD GVIDNPNVEL 60
61 VNRNLWSTFL ECGTEMIITK KGRRMFPLVK LKLSGLDKNS NYTIIMEMIS VDKLRYKFWN 120
121 GNWIVAGVGE HHPLPTCFVH PQSPRSGEWW MTDGVDFKMA KLSNNPFNND GHIVLNSMHR 180
181 YNPRFHIVRA DSSGQPILAS LKTFSFKETE FIAVTAYQND VVTKCKIDNN PFAKGFRNID 240
241 TVRKRKMNQI ISTPPASEDE EPEVKRTKSE IEAVMFSDPK VPQMSLQLIS QWQEALLSTI 300
301 VQIPITNQKS SSERKSGFGV VDLLGSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
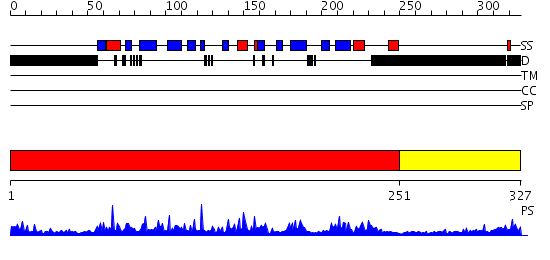
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..250] | 80.69897 | T-box protein 3, tbx3 | |
| 2 | View Details | [251..327] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.59 |
Source: Reynolds et al. (2008)