













| General Information: |
|
| Name(s) found: |
CE30601
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 618 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum
[IEA |
| Biological Process: |
positive regulation of growth rate
[IMP embryonic development ending in birth or egg hatching [IMP glycerol ether metabolic process [IEA cell redox homeostasis [IEA gamete generation [IMP positive regulation of multicellular organism growth [IMP |
| Molecular Function: |
protein disulfide oxidoreductase activity
[IEA electron carrier activity [IEA isomerase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMFDRRFFAL VVLLCVSAVR STEDASDDEL NYEMDEGVVV LTDKNFDAFL KKNPSVLVKF 60
61 YAPWCGHCKH LAPEYEKASS KVSIPLAKVD ATVETELGKR FEIQGYPTLK FWKDGKGPND 120
121 YDGGRDEAGI VEWVESRVDP NYKPPPEEVV TLTTENFDDF ISNNELVLVE FYAPWCGHCK 180
181 KLAPEYEKAA QKLKAQGSKV KLGKVDATIE KDLGTKYGVS GYPTMKIIRN GRRFDYNGPR 240
241 EAAGIIKYMT DQSKPAAKKL PKLKDVERFM SKDDVTIIGF FATEDSTAFE AFSDSAEMLR 300
301 EEFKTMGHTS DPAAFKKWDA KPNDIIIFYP SLFHSKFEPK SRTYNKAAAT SEDLLAFFRE 360
361 HSAPLVGKMT KKNAATRYTK KPLVVVYYNA DFSVQYREGS EYWRSKVLNI AQKYQKDKYK 420
421 FAVADEEEFA KELEELGLGD SGLEHNVVVF GYDGKKYPMN PDEFDGELDE NLEAFMKQIS 480
481 SGKAKAHVKS APAPKDDKGP VKTVVGSNFD KIVNDESKDV LIEFYAPWCG HCKSFESKYV 540
541 ELAQALKKTQ PNVVLAKMDA TINDAPSQFA VEGFPTIYFA PAGKKSEPIK YSGNRDLEDL 600
601 KKFMTKHGVK SFQKKDEL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
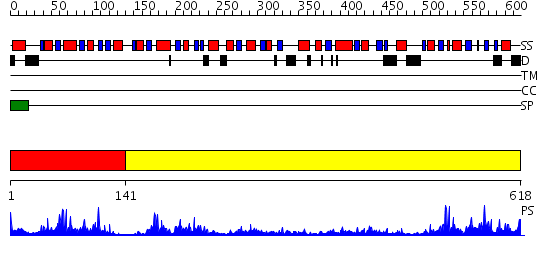
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..140] | 29.221849 | HUMAN PROTEIN DISULFIDE ISOMERASE, NMR, 40 STRUCTURES | |
| 2 | View Details | [141..618] | 69.39794 | Crystal Structure of Yeast Protein Disulfide Isomerase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|