













| General Information: |
|
| Name(s) found: |
akt-2 /
CE29298
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 483 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IDA |
| Biological Process: |
dauer larval development
[IMP insulin receptor signaling pathway [IMP determination of adult lifespan [IMP protein amino acid phosphorylation [IEA |
| Molecular Function: |
ATP binding
[IEA protein tyrosine kinase activity [IEA zinc ion binding [IEA protein kinase activity [IEA protein serine/threonine kinase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTENAHLQK EDIVIESWLH KKGEHIRNWR PRYFILFRDG TLLGFRSKPK EDQPLPEPLN 60
61 NFMIRDAATV CLDKPRPNMF IVRCLQWTTV IERTFYADSA DFRQMWIEAI QAVSSHNRLK 120
121 ENAGNTSMQE EDTNGNPSGE SDVNMDATST RSDNDFESTV MNIDEPEEVP RKNTVTMDDF 180
181 DFLKVLGQGT FGKVILCREK SSDKLYAIKI IRKEMVVDRS EVAHTLTENR VLYACVHPFL 240
241 TLLKYSFQAQ YHICFVMEFA NGGELFTHLQ RCKTFSEART RFYGSEIILA LGYLHHRNIV 300
301 YRDMKLENLL LDRDGHIKIT DFGLCKEEIK YGDKTSTFCG TPEYLAPEVI EDIDYDRSVD 360
361 WWGVGVVMYE MMCGRLPFSA KENGKLFELI TTCDLKFPNR LSPEAVTLLS GLLERVPAKR 420
421 LGAGPDDARE VSRAEFFKDV DWEATLRKEV EPPFKPNVMS ETDTSFFDRV RYVSILLKVS 480
481 EAI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
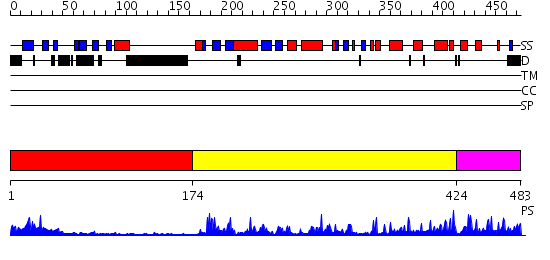
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..173] | 21.522879 | Solution Structure of the Pleckstrin Homology Domain of Human Protein Kinase B beta (Pkb/Akt) | |
| 2 | View Details | [174..423] | 53.39794 | Crystal Structure Of The Human P21-Activated Kinase 6 | |
| 3 | View Details | [424..483] | 2.36 | Structure of cAMP-dependent protein kinase complexed with Isoquinoline-5-sulfonic acid (2-(2-(4-chlorobenzyloxy)ethylamino) ethyl)amide |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)