













| General Information: |
|
| Name(s) found: |
rpn-10 /
CE26355
[WormBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 346 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
spermatogenesis
[IGI ubiquitin-dependent protein catabolic process [IGI |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVQESTMICV DNSEYMRNGD FQPTRLQSQQ DAVNLVTQCK LRANPENAVG ILSMANSVQV 60
61 LSSLSTEQGR LMMKNHSIEP FGKCNFIAGI KIAHLALKHR QNRNHKMRVV LFIGSPLEEI 120
121 EMNELVKIAK KMKKEKVLCD VIMFGENESD GHEKFSTFVD TLNGKEGSGS SLIVVPQGSS 180
181 LTDALLQSSV CKNEDGQAAF GGGGNGMDNA FGMDVENDPD LALALRVSME EERARQAAAA 240
241 AANGGAADSG ADAEVAAAAA AVPLEEMDMG AMTEEQQLEW ALRLSMQENA PAEQPQVQHE 300
301 QMDVDGAPAV GGDNLDDLMN NPELLQQIVD DLPAANAEKD DDKEKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
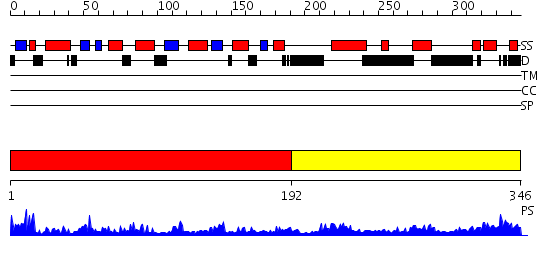
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..191] | 21.0 | Integrin alpha-x beta2 | |
| 2 | View Details | [192..346] | 13.69897 | Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)