













| General Information: |
|
| Name(s) found: |
air1 /
SPBP35G2.08c
[Sanger Pombe]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 313 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS nucleolus [ISS TRAMP complex [IDA |
| Biological Process: |
mRNA export from nucleus
[ISS rRNA catabolic process [ISS snRNA catabolic process [ISS snoRNA catabolic process [ISS tRNA catabolic process [ISS ncRNA polyadenylation involved in polyadenylation-dependent ncRNA catabolic process [ISS |
| Molecular Function: |
zinc ion binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTVSNQKAES DDGSDIDDAA LLQKINSLPI DQSITNSVSL EKHDFQGSDD HDSSTDLSDS 60
61 TLEDVEGSEW ADVSRGRYFG SDPSESIVCH NCKGNGHISK DCPHVLCTTC GAIDDHISVR 120
121 CPWTKKCMNC GLLGHIAARC SEPRKRGPRV CRTCHTDTHT SSTCPLIWRY YVEKEHPVRI 180
181 DVSEVRKFCY NCASDEHFGD DCTLPSRSNY PESTAFCEAN CPSGNDASNK EFFETRRKEF 240
241 QLERREQNRN QKSSKQRPFH KPGNSLASRL GSKPKSFKRK HSPPSEENGN LSFHSSDGRK 300
301 FTKTSKKNRK RKW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
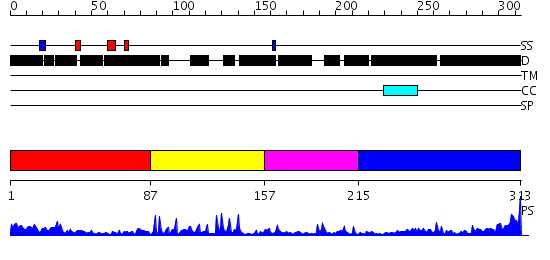
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..86] | 14.221849 | HIV-1 CAPSID PROTEIN C-TERMINAL FRAGMENT PLUS GAG P2 DOMAIN | |
| 2 | View Details | [87..156] | 5.39794 | NUCLEOCAPSID ZINC FINGERS DETECTED IN RETROVIRUSES: EXAFS STUDIES ON INTACT VIRUSES AND THE SOLUTION-STATE STRUCTURE OF THE NUCLEOCAPSID PROTEIN FROM HIV-1 | |
| 3 | View Details | [157..214] | 2.22 | Solution Structure of the Zinc-finger domain in LIN-28 | |
| 4 | View Details | [215..313] | 4.69897 | Solution Structure of the Human Immunodeficiency Virus Type 1 p6 Protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)