













| General Information: |
|
| Name(s) found: |
SPAC1296.06
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 558 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
NADP or NADPH binding
[ISS]
FMN binding [ISS] FAD binding [ISS] oxidoreductase activity [ISS] iron ion binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKNSHIYILY GSETGTAEGL AESLFRSLTR MGYDVLVNSM DDFNLENLLR PLQCVFICST 60
61 TGQGEMPLNM RERIITYRFN WASKKLDSRL RQLGAQSFSS RGEGDEQHPD GVEGVFAYWC 120
121 NHLYSQLAAI KTPSRPAFGE FDLLPPSFQI IIDESLGCKV KGFEDNNIVR HSRGKIEATL 180
181 VHNKRISNIK HWQDVRHLAF KIPNFERWKP GDVAVLYPWN DDMSVNSFIE CMGWESIKYS 240
241 PLIISSNVAE RKLPWFPNIL NVFNLVKYVL SIHSVPSRTF FEMASHFSNN KMHKERLQEF 300
301 SSYKNIDDYY DYTTRPRRTV LETLQEFKSV QIPIEYALDA FPVIRGRQYS IANRCDNSTG 360
361 ILELAVALVK YQTILKSPRQ GICSRWICDL HENTSFNIDI LPGFLNLSYQ SNKPLIMVGP 420
421 GTGVAPLRAL IQERIYNGLK ENLLFFGCRN KSMDFLFEKD WEKYTEEGTL KLFCAFSRDQ 480
481 EKKKYVQHSI QENGELVYNL LNEKDGMFFV SGSSGKMPSS VKDAIAGIVS KYSGCSISDG 540
541 YSFVTSLEKK NRYYQETW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
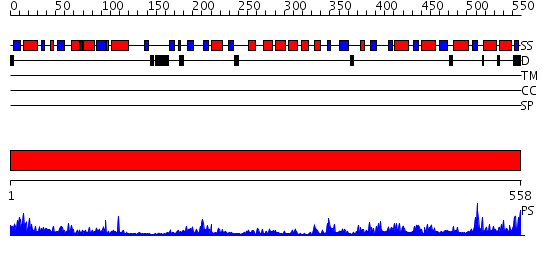
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..558] | 116.0 | NADPH-cytochrome p450 reductase; NADPH-cytochrome p450 reductase, N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)