













| General Information: |
|
| Name(s) found: |
mug76 /
SPCC1223.04c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 381 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA nucleolus [IDA |
| Biological Process: |
ribosome biogenesis
[TAS protein amino acid methylation [IDA |
| Molecular Function: |
protein-lysine N-methyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNKQNIESE VSWVKSKGAF VHPSLEFSVI PDAGSCVLAN NDINENTVLL KLPPNILINK 60
61 RTCSRYSFRD KLTSFQFLSW LISEDVHSNL EISPYYTKAL PQGFSFHPVT LTSDHPLWSI 120
121 LPDEVRNSLL ERKNVMAFDY EQVKKFVSVD QPTFQWGWLC VNTRCLYYDT GSKNTEDHLT 180
181 LAPIFEYFNH SPEAQTALIN TRGTITIKST RRIDKGEQIF LCYGPHGNDK LFTEYGFCLS 240
241 NNPNISIQLD RFIEFDKWQQ SFLQDHGYWN DYTCSLHGAS FRTLVGVRTL LVSPSEKLND 300
301 ASYDQTRRVL QYINGFSDGS RDRQDVEDYL KKVLQELLCE AEECKEKVKG ISDGSYVFIC 360
361 AEQLWKDRIM CCQYLMEHSF E |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
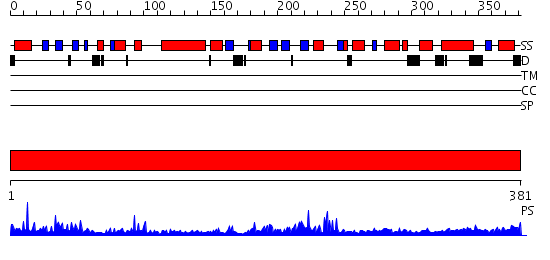
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..381] | 51.0 | RuBisCo LSMT C-terminal, substrate-binding domain; RuBisCo LSMT catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)