













| General Information: |
|
| Name(s) found: |
lsd2 /
SPAC23E2.02
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1273 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA Lsd1/2 complex [IDA nuclear chromatin [IC cytosol [IDA |
| Biological Process: |
chromatin remodeling
[ISS cellular response to heat [IEP negative regulation of transcription, DNA-dependent [IC positive regulation of transcription, DNA-dependent [IMP regulation of meiosis [IMP histone H3-K9 demethylation [IMP |
| Molecular Function: |
DNA binding
[IEA]
histone demethylase activity (H3-monomethyl-K4 specific) [NAS] transcription corepressor activity [ISS nucleosome binding [IDA histone demethylase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPLGRSSWIC CAKYFVNTKS RFNEILPPRF TLIVSFYSMN TSENDPDGHY DFPEMTEHHS 60
61 DRASSYANAN NVNSTQPQLV SSEQALLAIL GGGLQSITPN SVNQNAYSRS TYRTDGGLNS 120
121 QPVSSLNNQW GNYNPAFLPS RYDSSFHPYT ISQAANQPFP QHLLGNSNAG VAQQSGMRTI 180
181 GLPPTVGSSF PQQKSSTYEN FFDANSPSSQ QFPSTYPSRS QNPLSSSGDG STAIHAGPIQ 240
241 HQNSNAFSNY PYPLDASHLS SQQLLSMYRD QVSHGVTPST FRNHESFMPT QLVSATELSK 300
301 SVDNAVLPIP PTTAPAVVSP PASSFPLMSS AATSGNISSP ALFDSELGAR PEGSVAIEPS 360
361 RVLLQWSSQS SSHTIPSAGA SIPTSSLKSF FEHAAEAARK CNLDPRALES FEQHMLSDRL 420
421 HDPVVLFHYF QIRNSICWLW IKNPTHAISR VEAQGVCVDR CLFQLASLAY EFLVRYGYIN 480
481 YGCLSFDSSF TNETNTGTTS SSASKQKTIA VVGAGLTGLI CARQLTGLFS QYSSSFLSKN 540
541 ELPPKVIILE AKERTGGRIY SRALPVSHTS ATQINHHTSN SNSISSNSTS LNPKDVTDPS 600
601 HIPSAIDLGF QFLFSPMDDI LLNLLNKQLG IEVTEMTGSD LVYDETDTKV LDMVEVKKLN 660
661 ILWEKLLEYV SVCFFINVEE SVRISWISQF QLFIDEMFPD HLSKSLSLNA SHEFSFKKTM 720
721 LILIDEVSSY AKLGNSQKKF LIWCFKVAEL DDTLYPLNTV DTDFSKDILI PKVARRGLSQ 780
781 LPWALQSYPS PLNIHYEKFV SKVTIENDKC TLDCKDNSSY EVDQVVIACS PSHFSSNIEF 840
841 SPGLPNFVTE NIKSIDFKPG KKVILRYAAA FWRKNIRSFG IIPKSLSQEM NNDENDGKSC 900
901 FVLRIWNMLP ETGVPILVAD INPQMTSSSS NETSHLIQEL HSLIVDHFQN DSNSSADLLD 960
961 AWVTNWSRNG VYDGLNSYPN FANDKQQYEK RFRQSQLSYN LGRLHIAGDY IFSCVGCRTL1020
1021 QRSFLSGLSV CTGIIDSLAP ISLTIPIIGE TSRKELDQFL RNSKVNNFDP NAEAQRHLSY1080
1081 QARYRLKKQE RLDEHKEEQE QLVTELLGYL PEPPSKPNAN PFLLYQKMQW HVCRALADED1140
1141 KRRLTGDSTA KATINETRAK LGKTWRQLDD LGKKPWIDEI AAQREAYAGK ILRYQRLTKE1200
1201 YEMRAEQIRN DYAAKCQDEP IPDDEARLFM QAQREEEQRK QTQDDNISKS REASDEEYHD1260
1261 DGSSDSGYNG TRY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
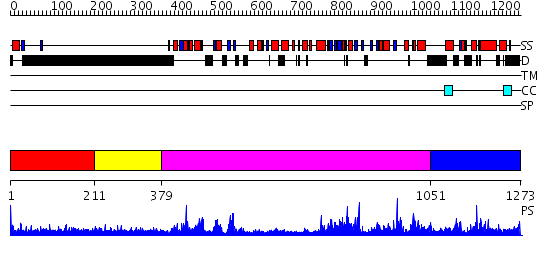
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..210] | 3.310996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [211..378] | 5.320996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [379..1050] | 160.0 | No description for 2v1dA was found. | |
| 4 | View Details | [1051..1273] | 23.045757 | No description for 2gzkA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)