













| General Information: |
|
| Name(s) found: |
SPAC31A2.06
[Sanger Pombe]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 542 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial inner membrane [ISS] |
| Biological Process: |
mitochondrial proton-transporting ATP synthase complex assembly
[ISS]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPTYRFNYLR FNKLCVSFFR SKFDKRPFAS QKFPENLVPD NSSNDANSQP EEVSSKKPWY 60
61 VDEKHNLFPK KAHFDAVALP PIPKGAPNFL ADVLNLLKKK YYATDLSFVN SPADSFWCDS 120
121 DLILLASCNC GSEVVSATNG LLRLLKQKNV GPVNVDGLTS ASRRKILERR MRKRSNLSNR 180
181 QLNTSENNWT CLSIENFGIS IHVITKNFRE YYKLDNIEHV KDETLYSDLE HGKQSRVSLT 240
241 SKSTPDNSLP PNFINNHSNV FRRSFHTCNF SLKSAASLYC DTQDILLNVN SQNLTSTLEK 300
301 YKKMHLQNPN NFSLDFTLSI FERLRKDSSL QLTTKDINTL FSTIALSPTK LSMASKHSKN 360
361 LVSERMLYLS LMYKSLVDLK TIDSFSLKLL FLKFMICSCM VKGESNFFLD NRIFLLERIM 420
421 NRYGIPMTID TFLLMQFILA KSNRWSEVWR RWDNLRKAGV VFNERLYNHV YLLAFESKNE 480
481 RVINYVLTNI FEDMVSQSPP IPASKLMATS LKKCVQSLPE KYAHSFPSVR NYIAKMENSM 540
541 TH |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
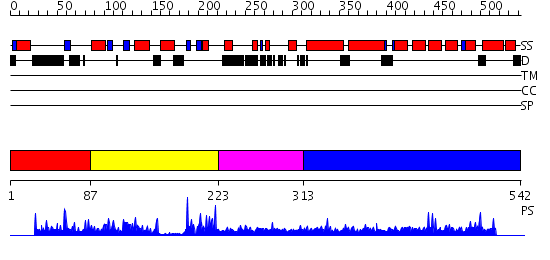
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..86] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [87..222] | 31.522879 | No description for 2o5aA was found. | |
| 3 | View Details | [223..312] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [313..542] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)