













| General Information: |
|
| Name(s) found: |
pst2 /
SPAC23C11.15
[Sanger Pombe]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1075 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA Rpd3S complex [IDA |
| Biological Process: |
chromatin remodeling
[IC chromosome condensation [IMP chromatin silencing [ISS histone deacetylation [IMP |
| Molecular Function: |
protein binding
[IPI transcription corepressor activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEQTLAILKN DNSTLVAEMQ NQLVHDFSPN GTALPELDIK AFVQKLGQRL CHRPYVYSAF 60
61 MDVVKALHNE IVDFPGFIER ISVILRDYPD LLEYLNIFLP SSYKYLLSNS GANFTLQFTT 120
121 PSGPVSTPST YVATYNDLPC TYHRAIGFVS RVRRALLSNP EQFFKLQDSL RKFKNSECSL 180
181 SELQTIVTSL LAEHPSLAHE FHNFLPSSIF FGSKPPLGSF PLRGIQSSQF TLSNISDLLS 240
241 QSRPDNLSPF SHLSNESSDF FKNVKNVLTD VETYHEFLKL LNLYVQGIID RNILVSRGFG 300
301 FLKSNSGLWR SFLSLTSLSP EEFLSVYNSA CSDFPECGPS YRLLPVEERN ISCSGRDDFA 360
361 WGILNDDWVS HPTWASEESG FIVQRKTPYE EAMTKLEEER YEFDRHIEAT SWTIKSLKKI 420
421 QNRINELPEE ERETYTLEEG LGLPSKSIYK KTIKLVYTSE HAEEMFKALE RMPCLTLPLV 480
481 ISRLEEKNEE WKSVKRSLQP GWRSIEFKNY DKSLDSQCVY FKARDKKNVS SKFLLAEADI 540
541 LRSQAKLHFP LRSRSAFEFS FVYDNEIVLF DTCYMVCTYI VCNSPSGLKK VEHFFKNILP 600
601 LHFGLEKDKF SIFLDQVFRG PDYDVNAPNI VGNKPVRRKR SNSITQLTEF VKQPKINGQR 660
661 ESRSAAAARK KEESGNKSQS NSQNSLSDES GNVTPVSKKQ LSQPAAAIKA SLKYPSHPDS 720
721 LLEHQDHAGD TENEMHDDVD KEQFGYSSMY VFFRLFNLLY ERLYELQRLE DQVSIIQQRI 780
781 IPNPVSQKQK IWRDRWNDLS DVPDEKTHYE NTYVMILRLI YGIVDQSAFE DYLRFYYGNK 840
841 AYKIYTIDKL VWSAAKQVHH IVSDGKYKFV TSLVEQNSSA SPKKNYDDFL YRLEIEKLLN 900
901 PDEILFRFCW INKFKSFGIK IMKRANLIVD QSLDTQRRVW KKYVQNYRIQ KLTEEISYKN 960
961 YRCPFLCRNI EKERTVEQLV SRLQTKLLRS AELVSGLQAK LCLDSFKLLY LPRTEDSYID1020
1021 ASYLRLRDTD FLDCQNKRKQ RWRNRWESLL KSVRGTSDNT AEVNFDADIN ALFIP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
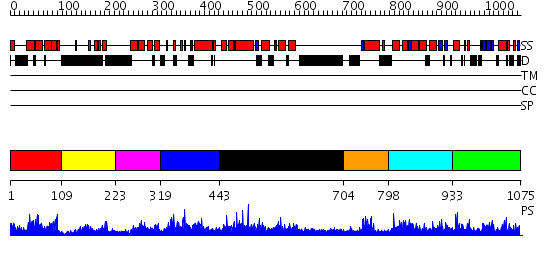
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..108] | 29.0 | Solution structure of the NRSF/REST-mSin3B PAH1 complex | |
| 2 | View Details | [109..222] | 18.39794 | No description for 2f05A was found. | |
| 3 | View Details | [223..318] | 2.81 | Solution structure of the first PAH domain of the mouse transcriptional repressor SIN3B | |
| 4 | View Details | [319..442] | 64.585027 | No description for PF08295.3 was found. No confident structure predictions are available. | |
| 5 | View Details | [443..703] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [704..797] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [798..932] | 1.033995 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [933..1075] | 2.032988 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)