













| General Information: |
|
| Name(s) found: |
arg11 /
SPAC4G9.09c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 885 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[IDA mitochondrion [IDA |
| Biological Process: |
arginine biosynthetic process
[IGI ornithine biosynthetic process [ISS phosphorylation [IC |
| Molecular Function: |
ATP binding
[IEA]
N-acetyl-gamma-glutamyl-phosphate reductase activity [IGI NAD or NADH binding [IEA] acetylglutamate kinase activity [IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLIELQQIVK SGLVRNGAKH CTKRSLLCSN ASVIASKRFQ GSFAPGQQQP LNPLAKPIEQ 60
61 DRDAIIRILS SIGSRREVEQ YLRYFTSFEA QRFAIIKVGG AIITDELDTL AQSLAFLNHV 120
121 GLYPIVVHGA GPQLNKILAS RNVEPEYSDG IRITDAETLS VARKVFLEEN AKLVDALEKL 180
181 GTRARPITGG VFQAEYLDKE KYKYVGKIVK VNKAPIEHSI RAGTLPILTS MAETASGQLL 240
241 NVNADITAGE LARVLKPLKV VYLNEKGGLI NGETKKKISS IYLDREYDGL MKQPWVKYGT 300
301 KLKIKEIKEL LDTLPRTSSV AIISTKDLQK ELFTESGAGT LISRGFVINK HDSLDSIPDA 360
361 ALENLIIQKN SLAAPSESLK QFKDTLKDRK LRIYSDSFNE SVAIVDTTDS SLPVLLAFGA 420
421 ADNNWLNNVV DSILTTLKAD FPSLLWRLQP SAKNLEWFFS KSEGTLFANN FYYFWYGVKD 480
481 LNKISKFIQS DKPFADAIIQ TQSTKPPTAS STTNNPSSSQ INQKRSYSTS SLFSKNKKMN 540
541 RSLFLKGGKR FFSAEAQKTQ KPLKAVSSKP AKVVLLGARG YTGKNLIGLI NTHPYLELSH 600
601 VSSRELEGTK LPGYTKKEIQ YVNLSTDDVK KLEEEGAVDA WVMALPNGVC KPYVDALTSA 660
661 NGKSVIVDLS ADYRFEPSWQ YGLPELNDRE ALRNSKRISN PGCYATGSQV GLTPILSLID 720
721 GQPSIFGVSG YSGAGTKPSP KNDLNVLTNN LIPYSLTDHI HEREISYRLK QPVAFIPHVA 780
781 QWFQGITLTI NVPLKKSITS RELRNLYQDR YNGEPLIHVQ GDVPLVKDNA HKHHVSVGGF 840
841 GVHSSGKRAV IVVTIDNLLK GAATQALQNL NLSCGYDEYA GIHLD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
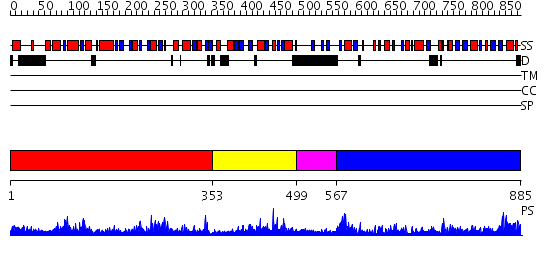
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..352] | 73.221849 | Acetylglutamate kinase from Thermotoga maritima complexed with its inhibitor arginine | |
| 2 | View Details | [353..498] | 14.522879 | No description for 2re1A was found. | |
| 3 | View Details | [499..566] | 1.99 | Crystal structure of putative N-acetyl-gamma-glutamyl-phosphate reductase (AK071544) from rice (Oryza sativa) | |
| 4 | View Details | [567..885] | 56.221849 | Crystal structure of N-acetyl-gamma-glutamyl-phosphate reductase (TM1782) from Thermotoga maritima at 1.80 A resolution |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)