













| General Information: |
|
| Name(s) found: |
meu7 /
SPBC16A3.13
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 774 amino acids |
Gene Ontology: |
|
| Cellular Component: |
external side of plasma membrane
[ISS endoplasmic reticulum [IDA cell surface [IC anchored to membrane [IEA] |
| Biological Process: |
fungal-type cell wall biogenesis
[ISS cell morphogenesis [ISS cellular polysaccharide catabolic process [IC extracellular polysaccharide metabolic process [IC] |
| Molecular Function: |
alpha-amylase activity
[ISS calcium ion binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKLSLKDLAL SIILVLGVAP PVQALSAEEW KKQSVYNVMT DRFATSKVVP HCDVKAEKYC 60
61 GGNWQGIVDH LDYIRDLGFT AISISPVVEQ LEGPEYHGEA YHGHRPKNYY RLNPHFGNEE 120
121 DLKELSDALH GKGMYLMVDV AINHTISEYF EDDYFKNTYF DKTHIDHKCK EHCSCHHDKF 180
181 PRPVPHNGTK PDHKPWKHEE HCHHGKFPRP VPHNGTKPDH KPWKHEEHCS CHHDKFPRPV 240
241 PHNGTKPDHK PWKHEEHCSC HHDKFPRPVP HNGTKPDHKP WKHEEHCHHG KFPRPIPHNG 300
301 TKPDHKPWKH EEHCHHGRFP RPVPHNGTKP DHKPWKHEEH CSCHHDKFSR PVPHNGTKPD 360
361 HKPWKHEEHC HHGKFLRPVP HNVTKPDHKP WKHEEHCHHG KFPRPVPHNG TKPDHKPWKH 420
421 EEHCSCHEDH SVHERPSAKG ALEDYIQTFV ETFQIDGIRF DAMGDKYRKY WPDFCKAAGV 480
481 FCMGDLKSSD SLKVCDWQND LEGLSNFPVR ESAVKAFLMN NPEGIVDLAD KMTDMRGLCV 540
541 NPLLLGNFVE NKDLPRMASI SSDPATLKNA IAFVLMGDGI PMMYNGQELA MRGEEVPYNR 600
601 PAIWEKGFPK RNPYYKFTST INRFRKAMIE SKDGEAFLNM QSDISIVDKR ILLLRRGKVI 660
661 TVLNNLGSYY IHFHYTVSAE EHKWMDVLSC KSVPVQDNRQ VFMVRNGEPM ILYPEESAYE 720
721 MGLCPSPIQL TEDYNSEGSL SINVEEAQTS KAPEQNRGFT NVLAALLLSL LMIL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
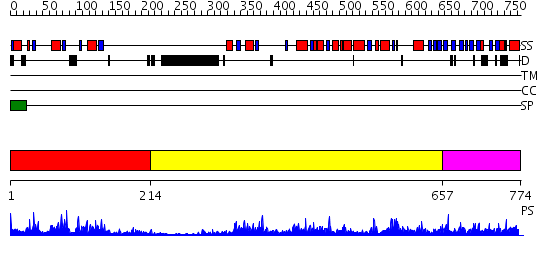
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..213] | 24.39794 | Crystal structure of Deinococcus radiodurans maltooligosyltrehalose trehalohydrolase | |
| 2 | View Details | [214..656] | 37.522879 | Glycosyltrehalose trehalohydrolase, N-terminal domain N; Glycosyltrehalose trehalohydrolase; Glycosyltrehalose trehalohydrolase, central domain | |
| 3 | View Details | [657..774] | 72.39794 | No description for 2guyA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|