













| General Information: |
|
| Name(s) found: |
Mes-4-PA /
FBpp0084636
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1427 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear chromosome
[ISS]
|
| Biological Process: |
gene silencing
[ISS]
germ-line sex determination [ISS] |
| Molecular Function: |
zinc ion binding
[IEA]
transcription cofactor activity [ISS] histone methyltransferase activity [IDA][ISS] histone-lysine N-methyltransferase activity [IEA] transcription factor binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKLSTDAHSE IEGDAAHGNV LCNSASDSLT ATDEVAAGND ESVATEGDDV EIPRDTNNST 60
61 PVRLLDKPGQ NPVQNGAQPA AEESELESQR QTPVQKQQQQ RVSMVNRKRD LINLQSALSP 120
121 KYIGYANANS PTPLSDSDDT IRTTRRRVNQ AAALNNSSAG ETLAHDNASP RTPGGGGGGG 180
181 GDDSANQLLS KTYMSPIEKL LIKNGASSPN STGFEAGSED LGIRPIVRKH VKRKMKRVPK 240
241 AKVTLELDEK NQQEVDEKSV KTEPIDEEVD RTDEAPTQEA QTTAISIKSE TEAEHKAAVD 300
301 VHIKQEDTIR LDIVNNPVES TSIVITEEPK DLEKSTEELA FALPLASSTE VDLKSPPDLS 360
361 STALATSIKS PSSVSIDSAK GLSIVTDPGW PTYQVGDLFW GKVFSYCFWP CMVCPDPLGQ 420
421 IVGNMPSHPQ RSSLDNANVP IQVHVRFFAD NGRRNWIKPE NLLTFAGLKA FDDMREELRI 480
481 KHGPKSAKYR QMVPKRTKVV IWRQAIEEAQ AMTQIPYSDR LEKFYQTYEN VVTLNRQKRK 540
541 RTKYMMQDTS DVGSSLYDST DNLHNKQGTQ LLAVKRERSE SPFSPAFSPV KSKNEKRAKR 600
601 RKLSNGTEAD TGSNSMAVTP SQTETTVDSS AYENPEFRQL LSAVMEYVMM NRSDEKVEKV 660
661 LLSVVSNIWS LKQIQLRELE RDLASGEIEE PLGSSVVGRG SGVGTIKRLS NRLMTMMVRR 720
721 SMTPVVTPST TPAPSEPDRR LSEPPKTKKP VNRPIEEVIE DILQLDSKYL FRGLSREPIC 780
781 KYCYQAGSDL VRCSRTCSSW LHADCLERKV TGAPMPKIGS RKALVIPPTS KSPSPDEDHV 840
841 TADAKEVVAV GTSLVCHECN VGEPEGCVIC HQVESPAVPS TPRKEDSSSH TPIEDKLLTC 900
901 SQPMCGKRFH TSCCKYWPQA SSSKHSARCP RHVCHTCVSD DPSGKFQQLG SSKLAKCVRC 960
961 PATYHQLSKC IPAGTQMLNT TNIICPRHNI AKADAHVNVL WCYICVKGGE LVCCETCPIA1020
1021 VHAHCRNIPI KTNESYICEE CESGRLPLYG EIVWAKFNNF RWWPAIILPP TEVPSNILKK1080
1081 AHGENDFVVR FFGTHDHGWI SRRRVYLYIE GDTGDGHKTK SQLFRNYTTG VEEASRFLPI1140
1141 IKARRQEQDM ERQSGNKLHP PPYVKIKTNK AVPPLRFSQN LEDLSTCNCL PVDEHPCGPE1200
1201 AGCLNRMLFN ECNPEYCKAG SLCENRMFEQ RKSPRLEVVY MNERGFGLVN REPIAVGDFV1260
1261 IEYVGEVINH AEFQRRMEQK QRDRDENYYF LGVEKDFIID AGPKGNLARF MNHSCEPNCE1320
1321 TQKWTVNCIH RVGIFAIKDI PVNSELTFNY LWDDLMNNSK KACFCGAKRC SGEIGGKLKD1380
1381 DAVKAHAKLK QMRRAKASAV RIHVKPKKTP KVKHISADDE PMDAKDE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
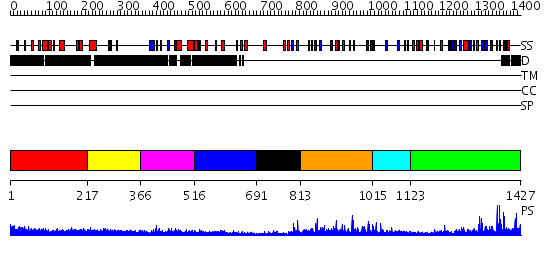
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..216] | 2.122996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [217..365] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [366..515] | 7.0 | No description for 2gfuA was found. | |
| 4 | View Details | [516..690] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [691..812] | 1.08 | Solution Structure of The C1 Domain of The Human Diacylglycerol Kinase Delta | |
| 6 | View Details | [813..1014] | 3.09691 | Type 1 insulin-like growth factor receptor extracellular domain; Type 1 insulin-like growth factor receptor Cys-rich domain | |
| 7 | View Details | [1015..1122] | 6.221849 | No description for 2daqA was found. | |
| 8 | View Details | [1123..1427] | 14.39794 | No description for 2igqA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)