













| General Information: |
|
| Name(s) found: |
noi-PA /
FBpp0078400
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 503 amino acids |
Gene Ontology: |
|
| Cellular Component: |
catalytic step 2 spliceosome
[IDA]
precatalytic spliceosome [IDA] spliceosomal complex [ISS] U2 snRNP [ISS] |
| Biological Process: |
positive regulation of NFAT protein import into nucleus
[IMP]
mitotic spindle organization [IMP] fertilization [IMP] nuclear mRNA splicing, via spliceosome [IC][ISS] |
| Molecular Function: |
nucleic acid binding
[IEA]
zinc ion binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METLLEQQRR LHEERERLVK LMVDEHATKK PGEKERIHSE HRLKYLMELH HNSTSQLRDL 60
61 YEDKDNERKA EIAALSGPNE FNEFYARLKQ IKQFYKSHPA EVSVPLSVEF DEMIRVYNNP 120
121 DDMSALVEFT DEEGGGRYLD LNECYELYLN LRSVEKLDYI TYLMSFDHVF DIPRERKNRE 180
181 YRIYIETLND YLHHFILRIQ PLLDLEGELL KVELDFQRQW LMGTFPGFSI KETESALANT 240
241 GAHLDLSAFS SWEELASLGL DRLKSALVAL GLKCGGTLEE RAQRLFSTKG KSTLDPALMA 300
301 KKPSAKTASA QSREHERHKE IAQLEALLYK YADLLSEQRA ATKENVQRKQ ARTGGERDDS 360
361 DVEASESDNE DDPDADDVPY NPKNLPLGWD GKPIPYWLYK LHGLNISYNC EICGNFTYKG 420
421 PKAFQRHFAE WRHAHGMRCL GIPNTAHFAN VTQIEDAITL WEKLKSQKQS ERWVADQEEE 480
481 FEDSLGNVVN RKTFEDLKRQ GLL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
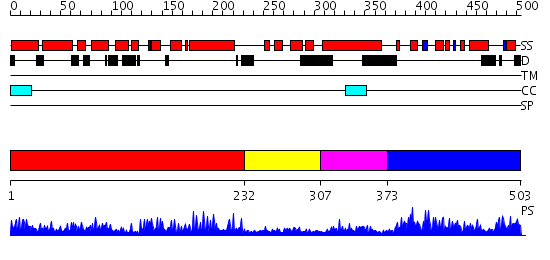
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..231] | 1.039947 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [232..306] | 7.051996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [307..372] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [373..503] | 1.05 | Solution Structure of a Human C2H2-type Zinc Finger Protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)