













| General Information: |
|
| Name(s) found: |
DhpD-PA /
FBpp0078625
[FlyBase]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 448 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
guanine metabolic process
[IMP]
eye pigment biosynthetic process [IMP] dihydropteridine metabolic process [IDA][IMP] |
| Molecular Function: |
zinc ion binding
[IEA]
guanine deaminase activity [IDA][ISS] dihydropterin deaminase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATVFLGTVV HTKSFSEFES FEGGFLAVDD AGKIIGVGQD YHAWASSNPA HAKGLTEVHL 60
61 SDYQFLMPGF VDCHIHAPQF AQLGLGLDMP LLDWLNTYTF PLEAKFSNHQ YAQQVYQGVV 120
121 EATLRCGTTL ASYFATNHLE STLTLAREAV RQGQRALIGK VCSNCNSPEF YVETAEESVS 180
181 ATLAFVEGVR KLGSPMVMPT ITPRFALSCS KELLKSLGDI AKRFDLHIQS HISENLEEIE 240
241 MVKGIFKTSY AGAYDEAGLL TNKTVLAHGV HLEDDEVALL KVRGCSVAHC PTSNTMLSSG 300
301 LCDVQRLVSG GVSVGLGTDV SGGNSVSIQD VLLRALDVSK HLDFFKKQNI RGTGVSKTQD 360
361 FNYHQLKYKQ ALYLATLGGA KALSLDHLTG NFALGKDFDA LLVDVSVVDK PLRRLSVDEL 420
421 VEKFIYTGSD RNIVEVFVAG KRIKQGYQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
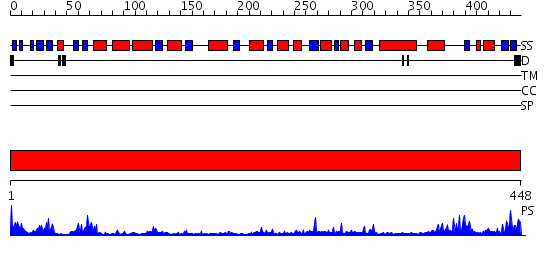
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..448] | 62.221849 | No description for 2uz9A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)