













| General Information: |
|
| Name(s) found: |
Klp59C-PA /
FBpp0071885
[FlyBase]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 626 amino acids |
Gene Ontology: |
|
| Cellular Component: |
kinetochore microtubule
[IDA]
kinesin complex [ISS] |
| Biological Process: |
mitotic sister chromatid segregation
[IMP]
microtubule-based movement [ISS] |
| Molecular Function: |
ATP binding
[IEA]
microtubule motor activity [ISS] motor activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDKLSIEQKI FIRRSDGRVH LAEIIKLEGG DSKLITVEWP EGHTVRGKEL PLELVVLMNP 60
61 HIFDSPRCSG GNAASANQTA SISPRSMKQR IATGSLSPVL ATAPPRQQTA PPVREDEVVH 120
121 QAERMRKERE RRREAQARTR LDREQGKNED PGNPNWEVAR MIRLQREQME SQRVRSGTTN 180
181 ERINCHQIMV CVRKRPLRRK ELADREQDVV SIPSKHTLVV HEPRKHVNLV KFLENHSFRF 240
241 DYVFDEECSN ATVYEFTARP LIKHIFDGGM ATCFAYGQTG SGKTYTMGGQ FPGRHQSSMD 300
301 GIYAMAAKDV FSTLKTVPYN KLNLKVYCSF FEIYGTRVFD LLMPGKPQLR VLEDRNQQVQ 360
361 VVGLTQNPVQ NTAEVLDLLE LGNSVRTSGH TSANSKSSRS HAVFQIVLRS AAGEKLHGKF 420
421 SLIDLAGNER GADNSSADRQ TRLEGSEINK SLLVLKECIR ALGRQSSHLP FRGSKLTQVL 480
481 RDSFIGGKKV KTCMIAMISP CLHSVEHTLN TLRYADRVKE LSVESIPSKR MPDANLGSTS 540
541 MSDIVCQSST QRLFPCASST SMPGGGNQAQ QHTNTANDLN RSQKPTSKPT YPTSGQQLVQ 600
601 RKGSSQREAS MMLTKSLAQF RGRNFP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
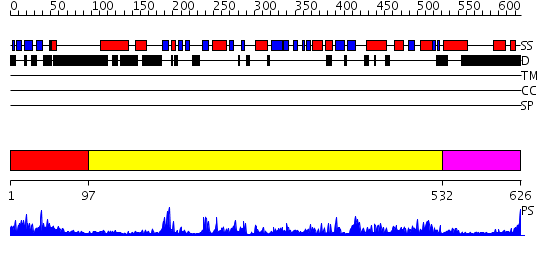
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..96] | 1.02 | Solution Structure of the Tudor Domain from Mouse Hypothetical Protein Homologous to Histone Acetyltransferase | |
| 2 | View Details | [97..531] | 105.0 | No description for 2gryA was found. | |
| 3 | View Details | [532..626] | 3.522879 | No description for 2dfsA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)