













| General Information: |
|
| Name(s) found: |
fus-PE /
FBpp0086479
[FlyBase]
fus-PD / FBpp0086477 [FlyBase] fus-PF / FBpp0086476 [FlyBase] |
| Description(s) found:
Found 49 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 967 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
epidermal growth factor receptor signaling pathway
[IGI]
|
| Molecular Function: |
nucleic acid binding
[IEA]
mRNA binding [ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQVPEHVVSL YIATCGQNGS GLGSDEKEII LLVFVLLEVS TGQIVGTKQI LVRPDGYFIK 60
61 DRTISSSSDN SSVTNNTASS PPLAIGDANN GSGSTSGNGN SLENGSELIL PIAEAQAAGK 120
121 PLSEAIEEFD AYLRSLSLHD TEIKLVTDGQ LPLRQCLHRE ASAKDVELPA YYNRFSDLRK 180
181 EFLRYKSGDL ARALVPVKDV KKMLQAPTLP MPQSIAEMLG ELNISSVEDN DFYIRESRDM 240
241 VTVIQTLLQA GHKFAANELV NLVLEPGICS IDDEVDGNCI VRARGLPWQS SDQDIAKFFR 300
301 GLNVAKGGVA LCLSPLGRRN GEALIRFVCQ EHRDMALKRH KHHIGTRYIE VYRASGEDFL 360
361 AIAGGASNEA QAFLSKGAQV IIRMRGLPYD ATAKQVLDFF TTGDTPPCHV LDGNEGVLFV 420
421 KKPDGRATGD AFVLFANETD APKALGRHRE SIGQRYIELF RSTTAEVQQV LNRSMDPKNY 480
481 ESGGGHSQPP LIAQLPTMQL PLLPQVGAHS LSHSLGASPA NLCPPVPPPA LPLPTQHLIT 540
541 SGTTKNCIRL RGLPYEAMVE HILHFLDDFA KHIIYQGVHM VINAQGQPSG EAFIQMDLEE 600
601 SARLCAQRRH NHYMMFGKKY RYIEVFQCSG DDMNMVLNGG LASPVAQPPP PHLGHAHKQQ 660
661 SLLAATTGMF SSAGQSPTTV AAGTAQSPLG GTHTHTHPHS HAHAHATGHA HAAAAAAHHG 720
721 LSASSAMLPG LSAAASASGL ASFMSAGQQS AAAAAHSAAS LQNAAVALAP QGYALNPFSL 780
781 PPPGSAAAAA ASTPALLAQQ QAQFIAQQSL LVRQQAAAAA LAAEQQQQQQ QQQQLYASAM 840
841 LQQHPLYLQQ QQQQQQLYAS AMLQQGQPQF VLMQRPSAAY LPPFPLSYMS AAGAASGVGV 900
901 APGAAVAGAA AAASPSASNS SLSQSMKRSY ENAFQQEAAG AAAAASAAKR ALTRQPSSVY 960
961 SYYNSGI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
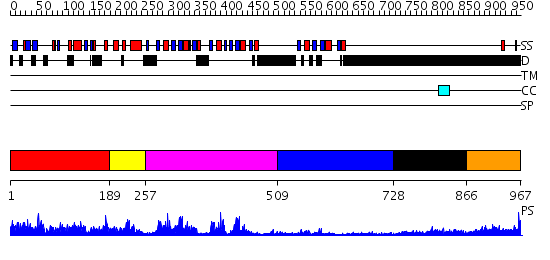
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..188] | 8.522879 | Crystallographic structure of the nuclease domain of 3'hExo, a DEDDh family member, bound to rAMP | |
| 2 | View Details | [189..256] | 28.69897 | No description for 2dhsA was found. | |
| 3 | View Details | [257..508] | 55.154902 | Nuclear ribonucleoprotein A1 (RNP A1, UP1) | |
| 4 | View Details | [509..727] | 3.88 | Nuclear ribonucleoprotein A1 (RNP A1, UP1) | |
| 5 | View Details | [728..865] | 7.0 | Sec24 | |
| 6 | View Details | [866..967] | 15.30103 | poly(A) binding protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 |
|
|||||||||||||||
| 4 |
|
|||||||||||||||
| 5 | No functions predicted. | |||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.26 |
Source: Reynolds et al. (2008)