













| General Information: |
|
| Name(s) found: |
PDAT_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 623 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASSKKSKTH KKKKEVKSPI DLPNSKKPTR ALSEQPSASE TQSVSNKSRK SKFGKRLNFI 60
61 LGAILGICGA FFFAVGDDNA VFDPATLDKF GNMLGSSDLF DDIKGYLSYN VFKDAPFTTD 120
121 KPSQSPSGNE VQVGLDMYNE GYRSDHPVIM VPGVISSGLE SWSFNNCSIP YFRKRLWGSW 180
181 SMLKAMFLDK QCWLEHLMLD KKTGLDPKGI KLRAAQGFEA ADFFITGYWI WSKVIENLAA 240
241 IGYEPNNMLS ASYDWRLSYA NLEERDKYFS KLKMFIEYSN IVHKKKVVLI SHSMGSQVTY 300
301 YFFKWVEAEG YGNGGPTWVN DHIEAFINIS GSLIGAPKTV AALLSGEMKD TGIVITLNIL 360
361 EKFFSRSERA MMVRTMGGVS SMLPKGGDVA PDDLNQTNFS NGAIIRYRED IDKDHDEFDI 420
421 DDALQFLKNV TDDDFKVMLA KNYSHGLAWT EKEVLKNNEM PSKWINPLET SLPYAPDMKI 480
481 YCVHGVGKPT ERGYYYTNNP EGQPVIDSSV NDGTKVENGI VMDDGDGTLP ILALGLVCNK 540
541 VWQTKRFNPA NTSITNYEIK HEPAAFDLRG GPRSAEHVDI LGHSELNEII LKVSSGHGDS 600
601 VPNRYISDIQ EIINEINLDK PRN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
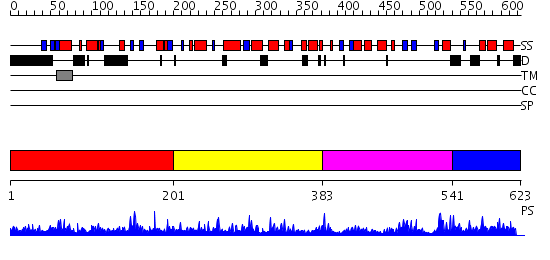
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..200] | 1.095966 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [201..382] | 22.045757 | Lipase | |
| 3 | View Details | [383..540] | 1.13498 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [541..623] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|