













| General Information: |
|
| Name(s) found: |
HLDE_YERPN
[Swiss-Prot]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Yersinia pestis Nepal516 |
| Length: | 476 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKVTLPDFRR AGVLVVGDVM LDRYWYGPTC RISPEAPVPV VKVDTIEERP GGAANVAMNI 60
61 ASLGAVARLV GLTGIDDAAR ALICKLSEVR VRCDFVSVPT HPTITKLRVL SRNQQLIRLD 120
121 FEEGFDGVDP TPIFERIQLA LPQIGALVLS DYAKGALNSV QPMIQLARKA NVPVLIDPKG 180
181 SDFERYRGAT LLTPNLSEFE AVVGRCKNEE ELVNRGMQLV ADFELSALLV TRSEQGMTLL 240
241 QLGKPPLHLP TQAKEVFDVT GAGDTVIGVL AAALAAGNSL EESCFLANAA AGVVVGKLGT 300
301 STVSPIELEN AIRGRAETGF GVMDEQQLKI AVAQARQRGE KVVMTNGIFD ILHAGHVSYL 360
361 ANARKLGDRL IVAVNSDAST KRLKGEKRPV NPLEQRMVVL GALEAVDWVV PFEEDTPQRL 420
421 IADILPDLLV KGGDYKPHEI AGSEEVWAAG GEVKVLNFED GVSTTNIIQS IKNGRG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
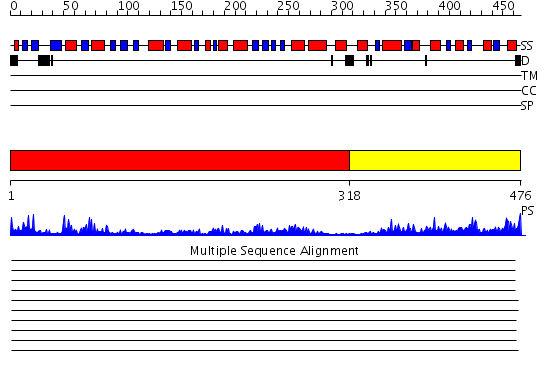
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..317] | 48.154902 | Ribokinase | |
| 2 | View Details | [318..476] | 28.69897 | CTP:glycerol-3-phosphate cytidylyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)