













| General Information: |
|
| Name(s) found: |
ADARB2
[HGNC (HUGO)]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 739 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASVLGSGRG SGGLSSQLKC KSKRRRRRRS KRKDKVSILS TFLAPFKHLS PGITNTEDDD 60
61 TLSTSSAEVK ENRNVGNLAA RPPPSGDRAR GGAPGAKRKR PLEEGNGGHL CKLQLVWKKL 120
121 SWSVAPKNAL VQLHELRPGL QYRTVSQTGP VHAPVFAVAV EVNGLTFEGT GPTKKKAKMR 180
181 AAELALRSFV QFPNACQAHL AMGGGPGPGT DFTSDQADFP DTLFQEFEPP APRPGLAGGR 240
241 PGDAALLSAA YGRRRLLCRA LDLVGPTPAT PAAPGERNPV VLLNRLRAGL RYVCLAEPAE 300
301 RRARSFVMAV SVDGRTFEGS GRSKKLARGQ AAQAALQELF DIQMPGHAPG RARRTPMPQE 360
361 FADSISQLVT QKFREVTTDL TPMHARHKAL AGIVMTKGLD ARQAQVVALS SGTKCISGEH 420
421 LSDQGLVVND CHAEVVARRA FLHFLYTQLE LHLSKRREDS ERSIFVRLKE GGYRLRENIL 480
481 FHLYVSTSPC GDARLHSPYE ITTDLHSSKH LVRKFRGHLR TKIESGEGTV PVRGPSAVQT 540
541 WDGVLLGEQL ITMSCTDKIA RWNVLGLQGA LLSHFVEPVY LQSIVVGSLH HTGHLARVMS 600
601 HRMEGVGQLP ASYRHNRPLL SGVSDAEARQ PGKSPPFSMN WVVGSADLEI INATTGRRSC 660
661 GGPSRLCKHV LSARWARLYG RLSTRTPSPG DTPSMYCEAK LGAHTYQSVK QQLFKAFQKA 720
721 GLGTWVRKPP EQQQFLLTL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
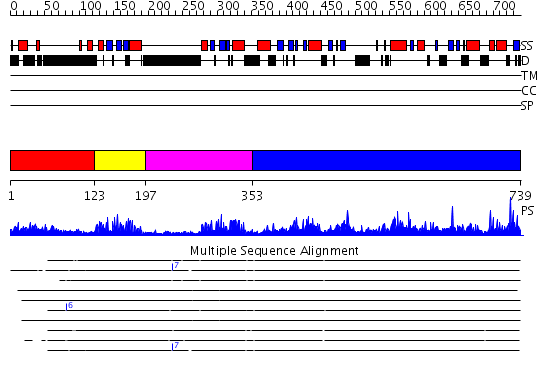
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..122] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [123..196] | 18.39794 | No description for 2b7tA was found. | |
| 3 | View Details | [197..352] | 18.154902 | No description for 2b7vA was found. | |
| 4 | View Details | [353..739] | 161.0 | Crystal structure of the catalytic domain of an adenosine deaminase that acts on RNA (hADAR2) bound to inositol hexakisphosphate (IHP) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|