













| General Information: |
|
| Name(s) found: |
MET22 /
YOL064C
[SGD]
|
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 357 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
methionine biosynthetic process
[TAS hyperosmotic salinity response [TAS response to drug [IMP sulfate assimilation [TAS |
| Molecular Function: |
3'(2'),5'-bisphosphate nucleotidase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALERELLVA TQAVRKASLL TKRIQSEVIS HKDSTTITKN DNSPVTTGDY AAQTIIINAI 60
61 KSNFPDDKVV GEESSSGLSD AFVSGILNEI KANDEVYNKN YKKDDFLFTN DQFPLKSLED 120
121 VRQIIDFGNY EGGRKGRFWC LDPIDGTKGF LRGEQFAVCL ALIVDGVVQL GCIGCPNLVL 180
181 SSYGAQDLKG HESFGYIFRA VRGLGAFYSP SSDAESWTKI HVRHLKDTKD MITLEGVEKG 240
241 HSSHDEQTAI KNKLNISKSL HLDSQAKYCL LALGLADVYL RLPIKLSYQE KIWDHAAGNV 300
301 IVHEAGGIHT DAMEDVPLDF GNGRTLATKG VIASSGPREL HDLVVSTSCD VIQSRNA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
MAK5 MET22 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
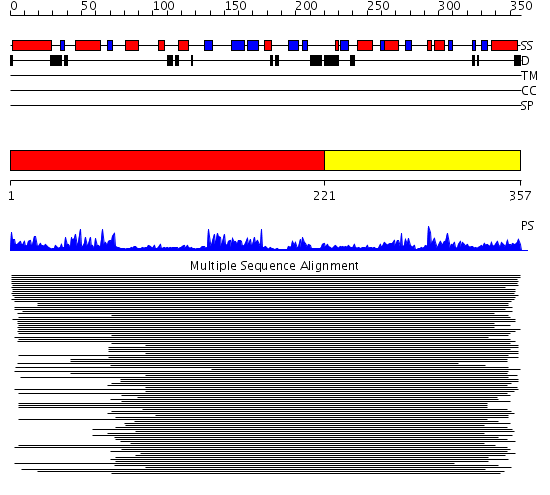
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..220] | 10085.69897 | 3';5'-adenosine bisphosphatase, PAP phosphatase | |
| 2 | View Details | [221..357] | 10085.69897 | 3';5'-adenosine bisphosphatase, PAP phosphatase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)